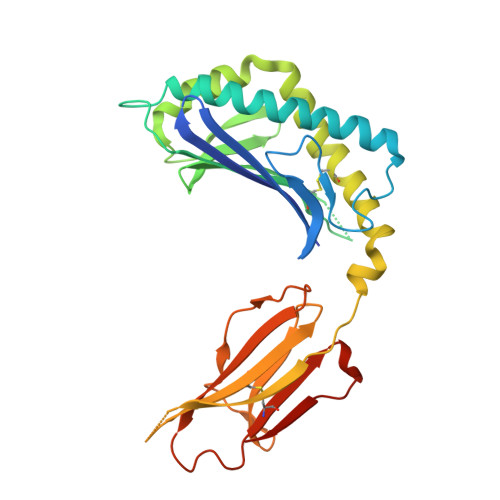
Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor.
Zajonc, D.M., Cantu, C., Mattner, J., Zhou, D., Savage, P.B., Bendelac, A., Wilson, I.A., Teyton, L.(2005) Nat Immunol 6: 810-818
- PubMed: 16007091
- DOI: https://doi.org/10.1038/ni1224
- Primary Citation of Related Structures:
1Z5L - PubMed Abstract:
Natural killer T cells express a conserved, semi-invariant alphabeta T cell receptor that has specificity for self glycosphingolipids and microbial cell wall alpha-glycuronosylceramide antigens presented by CD1d molecules. Here we report the crystal structure of CD1d in complex with a short-chain synthetic variant of alpha-galactosylceramide at a resolution of 2.2 A. This structure elucidates the basis for the high specificity of these microbial ligands and explains the restriction of the alpha-linkage as a unique pathogen-specific pattern-recognition motif. Comparison of the binding of altered lipid ligands to CD1d and T cell receptors suggested that the differential T helper type 1-like and T helper type 2-like properties of natural killer T cells may originate largely from differences in their 'loading' in different cell types and hence in their tissue distribution in vivo.
- Department of Molecular Biology and The Scripps Research Institute, La Jolla, California 92037, USA.
Organizational Affiliation: