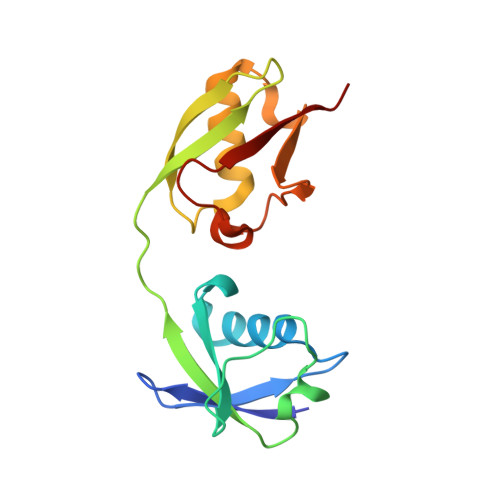
Crystal Structure of the Interferon-induced Ubiquitin-like Protein ISG15.
Narasimhan, J., Wang, M., Fu, Z., Klein, J.M., Haas, A.L., Kim, J.J.(2005) J Biological Chem 280: 27356-27365
- PubMed: 15917233
- DOI: https://doi.org/10.1074/jbc.M502814200
- Primary Citation of Related Structures:
1Z2M - PubMed Abstract:
The biological effects of the ISG15 protein arise in part from its conjugation to cellular targets as a primary response to interferon-alpha/beta induction and other markers of viral or parasitic infection. Recombinant full-length ISG15 has been produced for the first time in high yield by mutating Cys78 to stabilize the protein and by cloning in a C-terminal arginine cap to protect the C terminus against proteolytic inactivation. The cap is subsequently removed with carboxypeptidase B to yield mature biologically active ISG15 capable of stoichiometric ATP-dependent thiolester formation with its human UbE1L activating enzyme. The three-dimensional structure of recombinant ISG15C78S was determined at 2.4-A resolution. The ISG15 structure comprises two beta-grasp folds having main chain root mean square deviation (r.m.s.d.) values from ubiquitin of 1.7 A (N-terminal) and 1.0 A (C-terminal). The beta-grasp domains pack across two conserved 3(10) helices to bury 627 A2 that accounts for 7% of the total solvent-accessible surface area. The distribution of ISG15 surface charge forms a ridge of negative charge extending nearly the full-length of the molecule. Additionally, the N-terminal domain contains an apolar region comprising almost half its solvent accessible surface. The C-terminal domain of ISG15 was superimposed on the structure of Nedd8 (r.m.s.d. = 0.84 A) bound to its AppBp1-Uba3 activating enzyme to model ISG15 binding to UbE1L. The docking model predicts several key side-chain interactions that presumably define the specificity between the ubiquitin and ISG15 ligation pathways to maintain functional integrity of their signaling.
- Department of Biochemistry, Medical College of Wisconsin, Milwaukee, Wisconsin 53226, USA.
Organizational Affiliation: