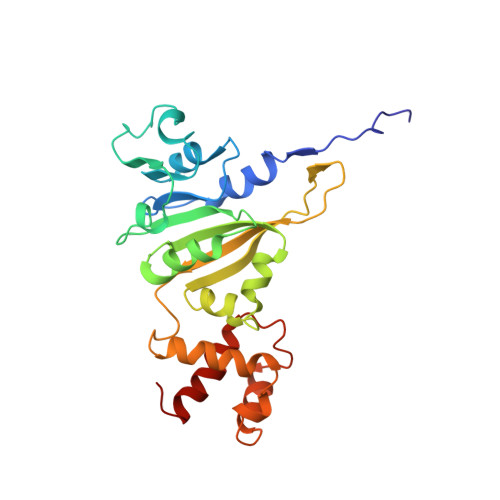
Solution structure of an rRNA methyltransferase (ErmAM) that confers macrolide-lincosamide-streptogramin antibiotic resistance.
Yu, L., Petros, A.M., Schnuchel, A., Zhong, P., Severin, J.M., Walter, K., Holzman, T.F., Fesik, S.W.(1997) Nat Struct Biol 4: 483-489
- PubMed: 9187657
- DOI: https://doi.org/10.1038/nsb0697-483
- Primary Citation of Related Structures:
1YUB - PubMed Abstract:
The Erm family of methyltransferases is responsible for the development of resistance to the macrolide-lincosamide-streptogramin type B (MLS) antibiotics. These enzymes methylate an adenine of 23S ribosomal RNA that prevents the MLS antibiotics from binding to the ribosome and exhibiting their antibacterial activity. Here we describe the three-dimensional structure of an Erm family member, ErmAM, as determined by NMR spectroscopy. The catalytic domain of ErmAM is structurally similar to that found in other methyltransferases and consists of a seven-stranded beta-sheet flanked by alpha-helices and a small two-stranded beta-sheet. In contrast to the catalytic domain, the substrate binding domain is different from other methyltransferases and adopts a novel fold that consists of four alpha-helices.
- Pharmaceutical Discovery Division, Abbott Laboratories, Abbott Park, Illinois 60064, USA.
Organizational Affiliation: