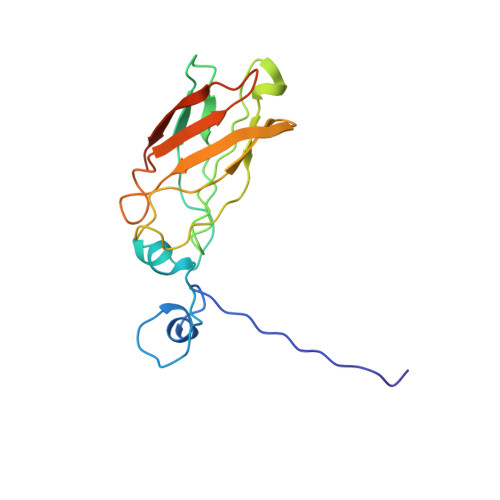
Solution structure of the immunodominant domain of protective antigen GNA1870 of Neisseria meningitidis
Cantini, F., Savino, S., Scarselli, M., Masignani, V., Pizza, M., Romagnoli, G., Swennen, E., Veggi, D., Banci, L., Rappuoli, R.(2006) J Biological Chem 281: 7220-7227
- PubMed: 16407174
- DOI: https://doi.org/10.1074/jbc.M508595200
- Primary Citation of Related Structures:
1YS5 - PubMed Abstract:
GNA1870, a 28-kDa surface-exposed lipoprotein of Neisseria meningitidis recently discovered by reverse vaccinology, is one of the most potent antigens of Meningococcus and a promising candidate for a universal vaccine against a devastating disease. Previous studies of epitope mapping and genetic characterization identified residues critical for bactericidal response within the C-terminal domain of the molecule. To elucidate the conformation of protective epitopes, we used NMR spectroscopy to obtain the solution structure of the immunodominant 18-kDa C-terminal portion of GNA1870. The structure consists of an eight-stranded antiparallel beta-barrel overlaid by a short alpha-helix with an unstructured N-terminal end. Residues previously shown to be important for antibody recognition were mapped on loops facing the same ridge of the molecule. The sequence similarity of GNA1870 with members of the bacterial transferrin receptor family allows one to predict the folding of this class of well known bacterial antigens, providing the basis for the rational engineering of high affinity B cell epitopes.
- Centro Risonanze Magnetiche (CERM), University of Florence, Via L. Sacconi 6, 50019 Sesto Fiorentino, Italy.
Organizational Affiliation: