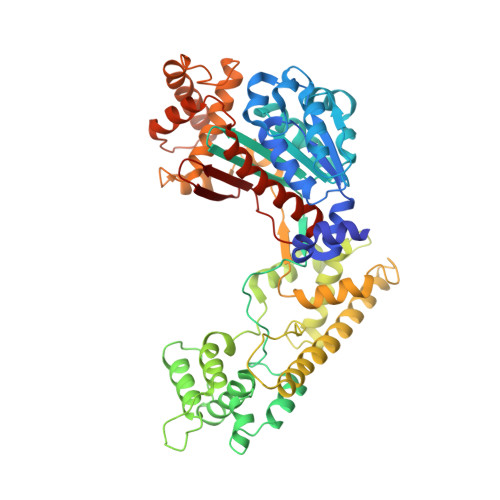
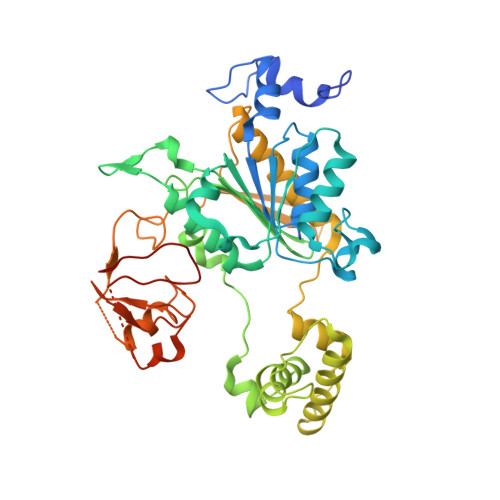
Insights into the Ubiquitin Transfer Cascade from the refined structure of the activating enzyme for NEDD8
Walden, H., Podgorski, M.S., Schulman, B.A.(2003) Nature 422: 330-334
- PubMed: 12646924
- DOI: https://doi.org/10.1038/nature01456
- Primary Citation of Related Structures:
1YOV - PubMed Abstract:
Post-translational modification by ubiquitin-like proteins (Ublps) is an essential cellular regulatory mechanism. The Ublp NEDD8 regulates cell division, signalling and embryogenesis. Ublps are conjugated to their targets by the sequential action of E1, E2 and often E3 enzymes. Each Ublp has a dedicated E1, or activating enzyme, that initiates its conjugation cascade. First, E1 associates with the Ublp and catalyses adenylation of the carboxy terminus of the Ublp. Second, E1 forms a thioester between its catalytic cysteine and the Ublp. Next, E1 is loaded with a second Ublp molecule, adenylating the C terminus of this second Ublp while still carrying the first thioester-bound Ublp. Last, E1 binds E2 and promotes Ublp transfer to the catalytic cysteine of E2. We report here the structure and mutational analysis of human APPBP1-UBA3, the heterodimeric E1 enzyme for NEDD8 (ref. 11). Each E1 activity is specified by a domain: an adenylation domain resembling bacterial adenylating enzymes, an E1-specific domain organized around the catalytic cysteine, and a domain involved in E2 recognition resembling ubiquitin. The domains are arranged around two clefts that coordinate protein and nucleotide binding so that each of E1's reactions drives the next, in an assembly-line fashion.
- Department of Structural Biology, St Jude Children's Research Hospital, Memphis, Tennessee 38105, USA.
Organizational Affiliation: