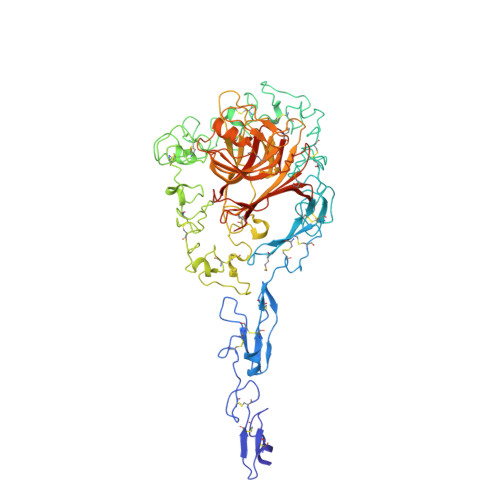
Structure of the calcium-rich signature domain of human thrombospondin-2
Carlson, C.B., Bernstein, D.A., Annis, D.S., Misenheimer, T.M., Hannah, B.L., Mosher, D.F., Keck, J.L.(2005) Nat Struct Mol Biol 12: 910-914
- PubMed: 16186819
- DOI: https://doi.org/10.1038/nsmb997
- Primary Citation of Related Structures:
1YO8 - PubMed Abstract:
Thrombospondins (THBSs) are secreted glycoproteins that have key roles in interactions between cells and the extracellular matrix. Here, we describe the 2.6-A-resolution crystal structure of the glycosylated signature domain of human THBS2, which includes three epidermal growth factor-like modules, 13 aspartate-rich repeats and a lectin-like module. These elements interact extensively to form three structural regions termed the stalk, wire and globe. The THBS2 signature domain is stabilized by these interactions and by a network of 30 bound Ca(2+) ions and 18 disulfide bonds. The structure suggests how genetic alterations of THBSs result in disease.
- Department of Medicine, 4285B Medical Sciences Center, University of Wisconsin-Madison, 1300 University Avenue, Madison, Wisconsin 53706, USA.
Organizational Affiliation: