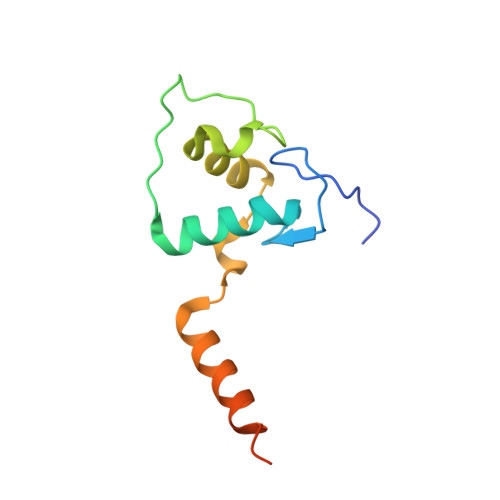
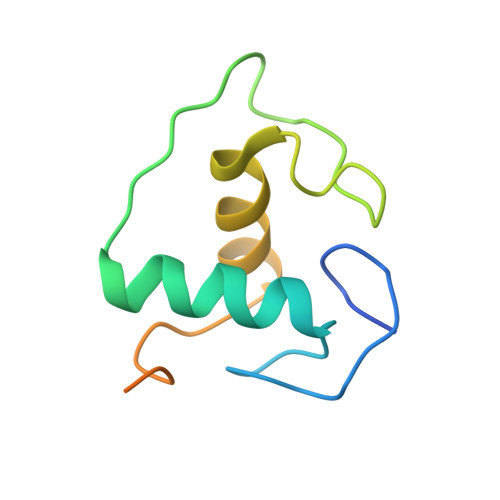
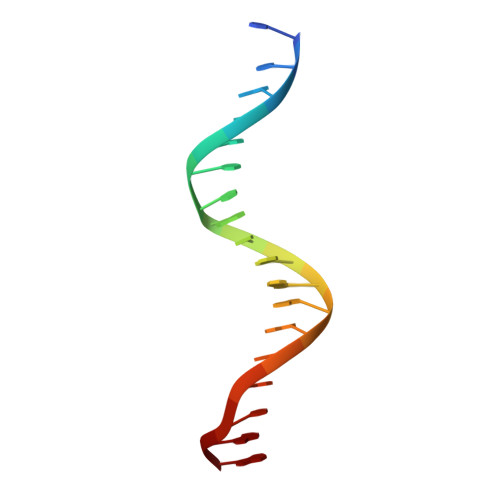
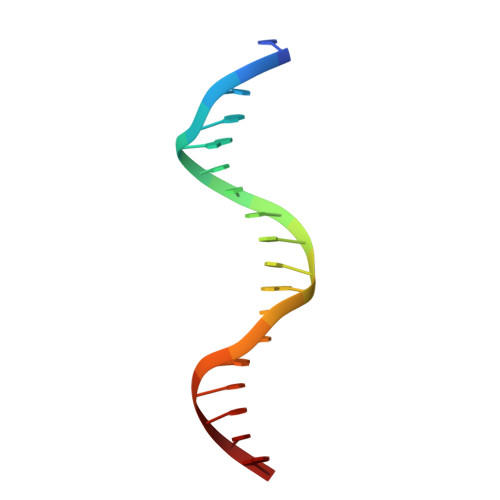
Structural analysis of RXR-VDR interactions on DR3 DNA
Shaffer, P.L., Gewirth, D.T.(2004) J Steroid Biochem Mol Biol 89-90: 215-219
- PubMed: 15225774
- DOI: https://doi.org/10.1016/j.jsbmb.2004.03.084
- Primary Citation of Related Structures:
1YNW - PubMed Abstract:
The Vitamin D receptor (VDR) is a ligand-responsive transcription factor that forms homo- or heterodimers on response elements composed of two hexameric half-sites separated by three base pairs of spacer DNA. Binding of 1alpha,25-dihydroxyvitamin D(3) to the full-length VDR causes destabilization of the VDR homodimer and formation of a heterodimeric complex with the 9-cis retinoic acid receptor (RXR). VDR and RXR DNA-binding domains (DBDs) do not mimic this behavior, however: VDR DBD homodimers are formed exclusively, even in the presence of excess RXR DBD. Exploiting the asymmetry of the heterodimer and our knowledge of the homodimeric DBD interface, we have engineered VDR mutants that disfavor the homodimeric complex and allow for the formation of heterodimeric DBD complexes with RXR on DR3 elements. One of these complexes has been crystallized and its structure determined. However, the polarity of the proteins relative to the DNA is non-physiological due to crystal packing between symmetry-related VDR DBD protomers. This reveals a flattened energy landscape that appears to rely on elements outside of the core DBD for response element discrimination in the heterodimer.
- Department of Biochemistry, Duke University Medical Center, Durham, NC 27710, USA.
Organizational Affiliation: