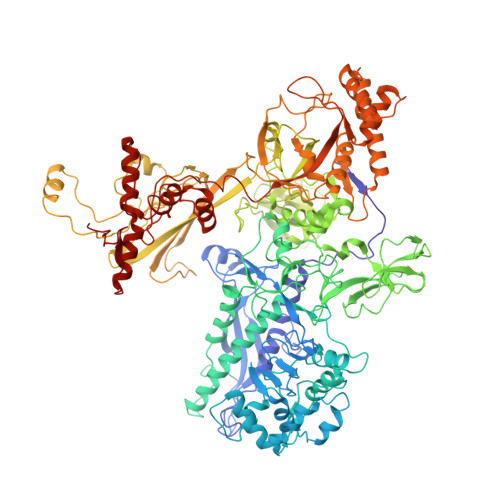
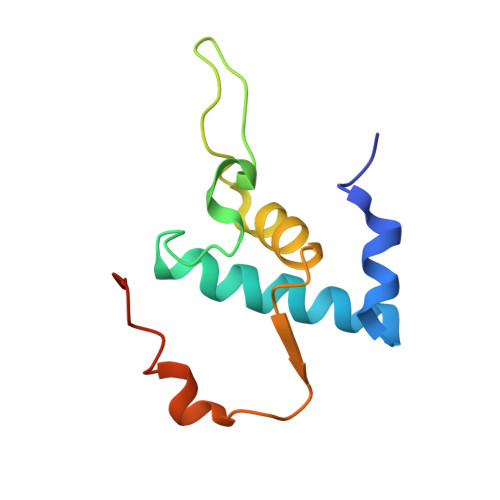
Structural, functional, and genetic analysis of sorangicin inhibition of bacterial RNA polymerase
Campbell, E.A., Pavlova, O., Zenkin, N., Leon, F., Irschik, H., Jansen, R., Severinov, K., Darst, S.A.(2005) EMBO J 24: 674-682
- PubMed: 15692574
- DOI: https://doi.org/10.1038/sj.emboj.7600499
- Primary Citation of Related Structures:
1YNJ, 1YNN - PubMed Abstract:
A combined structural, functional, and genetic approach was used to investigate inhibition of bacterial RNA polymerase (RNAP) by sorangicin (Sor), a macrolide polyether antibiotic. Sor lacks chemical and structural similarity to the ansamycin rifampicin (Rif), an RNAP inhibitor widely used to treat tuberculosis. Nevertheless, structural analysis revealed Sor binds in the same RNAP beta subunit pocket as Rif, with almost complete overlap of RNAP binding determinants, and functional analysis revealed that both antibiotics inhibit transcription by directly blocking the path of the elongating transcript at a length of 2-3 nucleotides. Genetic analysis indicates that Rif binding is extremely sensitive to mutations expected to change the shape of the antibiotic binding pocket, while Sor is not. We suggest that conformational flexibility of Sor, in contrast to the rigid conformation of Rif, allows Sor to adapt to changes in the binding pocket. This has important implications for drug design against rapidly mutating targets.
- The Rockefeller University, New York, NY 10021, USA.
Organizational Affiliation: