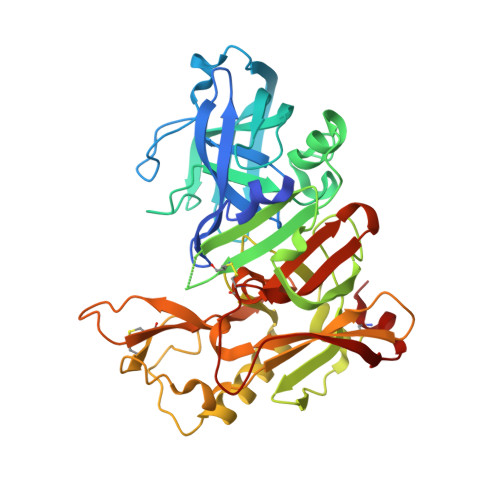
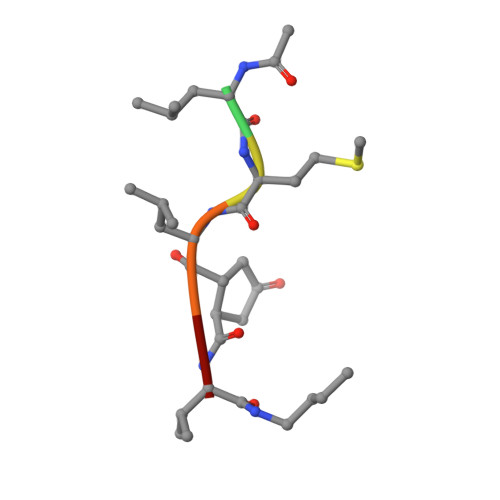
Structure-based design, synthesis, and memapsin 2 (BACE) inhibitory activity of carbocyclic and heterocyclic peptidomimetics
Hanessian, S., Yun, H., Hou, Y., Yang, G., Bayrakdarian, M., Therrien, E., Moitessier, N., Roggo, S., Veenstra, S., Tintelnot-Blomley, M., Rondeau, J.M., Ostermeier, C., Strauss, A., Ramage, P., Paganetti, P., Neumann, U., Betschart, C.(2005) J Med Chem 48: 5175-5190
- PubMed: 16078837
- DOI: https://doi.org/10.1021/jm050142+
- Primary Citation of Related Structures:
1YM2, 1YM4 - PubMed Abstract:
Molecular modeling based on the X-ray crystal structure of the Tang-Ghosh heptapeptide inhibitor 1 (OM99-2) of BACE led to the design and synthesis of a series of constrained P(1)' analogues. A cyclopentane ring was incorporated in 1 spanning the P(1)' Ala methyl group and the adjacent methylene carbon atom of the chain. Progressive truncation at the P(2)'-P(4)' sites led to a potent truncated analogue 5 with good selectivity over Cathepsin D. Using the same backbone replacement concept, a series of cyclopentane, cyclopentanone, tetrahydrofuran, pyrrolidine, and pyrrolidinone analogues were synthesized with considerable variation at the P and P' sites. The cyclopentanone and 2-pyrrolidinone analogues 45 and 57 showed low nM BACE inhibition. X-ray cocrystal structures of two analogues 5 and 45 revealed excellent convergence with the original inhibitor 1 structure while providing new insights into other interactions which could be exploited for future modifications.
- Department of Chemistry, Université de Montréal, C. P. 6128, Succursale Centre-Ville, Montréal, Province Quebec, Canada H3C 3J7. stephen.hanessian@umontreal.ca
Organizational Affiliation: