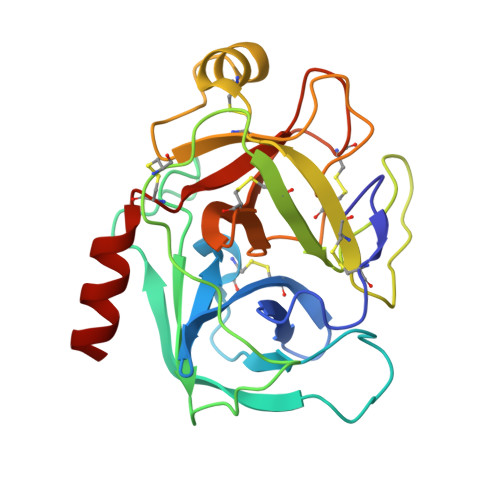
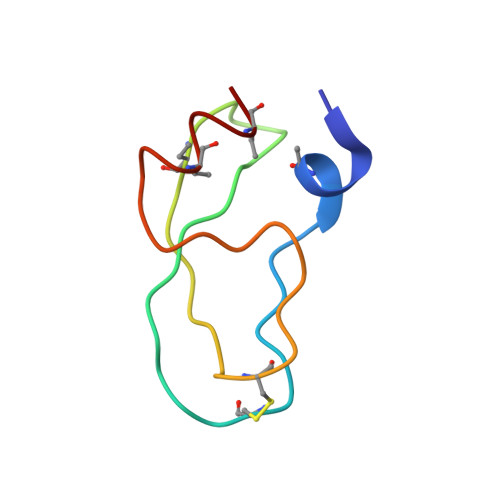
Partially folded bovine pancreatic trypsin inhibitor analogues attain fully native structures when co-crystallized with S195A rat trypsin
Getun, I.V., Brown, C.K., Tulla-Puche, J., Ohlendorf, D., Woodward, C., Barany, G.(2008) J Mol Biology 375: 812-823
- PubMed: 18054043
- DOI: https://doi.org/10.1016/j.jmb.2007.10.084
- Primary Citation of Related Structures:
1YKT, 1YLC, 1YLD - PubMed Abstract:
Crystal structures, at 1.7 A resolution, were solved for complexes between each of two chemically synthesized partially folded analogues of bovine pancreatic trypsin inhibitor (BPTI) with the proteolytically inactive rat trypsin mutant S195A. The BPTI analogue termed [14-38](Abu) retains only the disulfide bond between Cys14 and Cys38, while Cys5, Cys30, Cys51, and Cys55 are replaced by isosteric alpha-amino-n-butyric acid residues. The analogue K26P,A27D[14-38](Abu) contains two further replacements, by statistically favored residues, in the type I beta-turn that has been suggested to be a main site for initiation of BPTI folding. As a control, the structure of the complex between S195A trypsin and wild-type BPTI was also solved. Despite significant differences in the degree of structure detected among these three BPTIs in solution by several biophysical techniques, their tertiary folds once bound to S195A trypsin in a crystalline lattice are essentially superimposable.
- Department of Chemistry, University of Minnesota, Minneapolis, MN 55455, USA.
Organizational Affiliation: