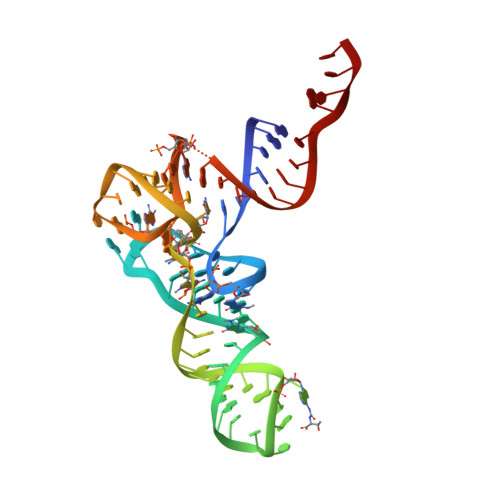
The 3 A crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination.
Basavappa, R., Sigler, P.B.(1991) EMBO J 10: 3105-3111
- PubMed: 1915284
- DOI: https://doi.org/10.1002/j.1460-2075.1991.tb07864.x
- Primary Citation of Related Structures:
1YFG - PubMed Abstract:
A significantly improved molecular model of yeast initiator tRNA (ytRNA(iMet) has been prepared that gives insight into the structural basis of eukaryotic initiator tRNA's unique function. This study was made possible by X-ray data collected at synchrotron radiation sources with the newly developed technologies of 'imaging plates' and 'storage phosphors'. These data extend beyond the resolution limit of 4.0 A reported previously to a current limit of 3.0 A and are considerably more accurate. Refinement of the model against the new data (R factor = 21.5%) clearly reveals a novel modification and a set of tertiary interactions involving sequence features characteristic of eukaryotic initiator tRNAs. We hypothesize these to be the structural elements responsible for part of the special function of yeast tRNA(iMET).
- Department of Biochemistry and Molecular Biology, University of Chicago.
Organizational Affiliation: