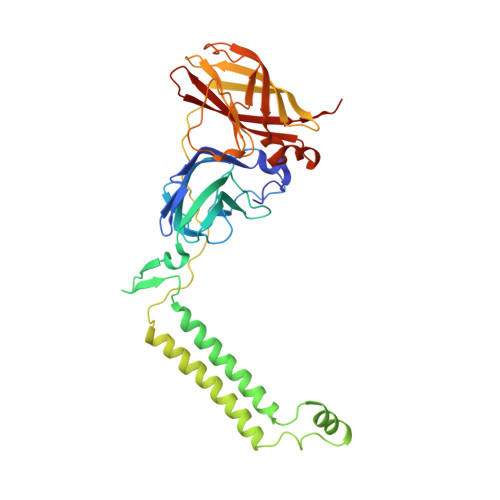
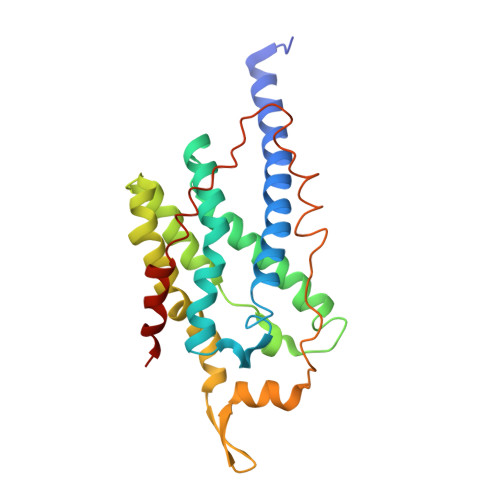
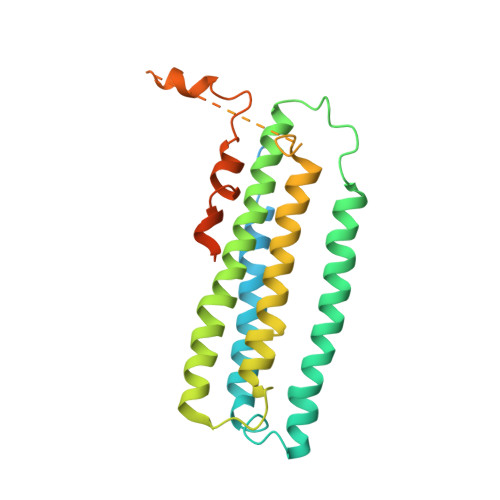
Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane.
Lieberman, R.L., Rosenzweig, A.C.(2005) Nature 434: 177-182
- PubMed: 15674245
- DOI: https://doi.org/10.1038/nature03311
- Primary Citation of Related Structures:
1YEW - PubMed Abstract:
Particulate methane monooxygenase (pMMO) is an integral membrane metalloenzyme that catalyses the conversion of methane to methanol. Knowledge of how pMMO performs this extremely challenging chemistry may have an impact on the use of methane as an alternative energy source by facilitating the development of new synthetic catalysts. We have determined the structure of pMMO from the methanotroph Methylococcus capsulatus (Bath) to a resolution of 2.8 A. The enzyme is a trimer with an alpha3beta3gamma3 polypeptide arrangement. Two metal centres, modelled as mononuclear copper and dinuclear copper, are located in soluble regions of each pmoB subunit, which resembles cytochrome c oxidase subunit II. A third metal centre, occupied by zinc in the crystal, is located within the membrane. The structure provides new insight into the molecular details of biological methane oxidation.
- Department of Biochemistry, Molecular Biology, and Cell Biology, Northwestern University, Evanston, Illinois 60208, USA.
Organizational Affiliation: