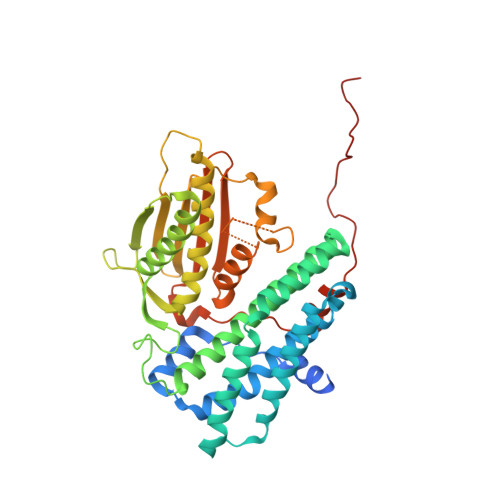
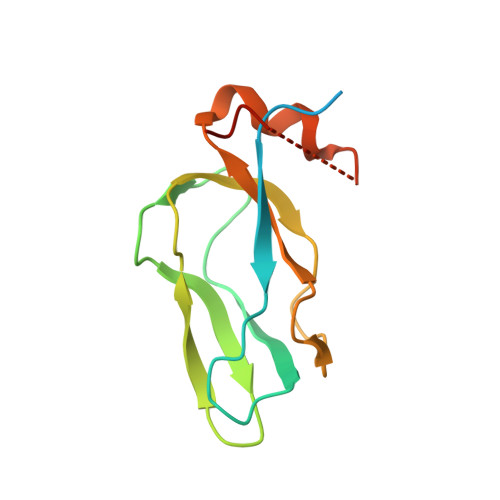
Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Kato, M., Chuang, J.L., Tso, S.C., Wynn, R.M., Chuang, D.T.(2005) EMBO J 24: 1763-1774
- PubMed: 15861126
- DOI: https://doi.org/10.1038/sj.emboj.7600663
- Primary Citation of Related Structures:
1Y8N, 1Y8O, 1Y8P - PubMed Abstract:
The human pyruvate dehydrogenase complex (PDC) is regulated by reversible phosphorylation by four isoforms of pyruvate dehydrogenase kinase (PDK). PDKs phosphorylate serine residues in the dehydrogenase (E1p) component of PDC, but their amino-acid sequences are unrelated to eukaryotic Ser/Thr/Tyr protein kinases. PDK3 binds to the inner lipoyl domains (L2) from the 60-meric transacetylase (E2p) core of PDC, with concomitant stimulated kinase activity. Here, we present crystal structures of the PDK3-L2 complex with and without bound ADP or ATP. These structures disclose that the C-terminal tail from one subunit of PDK3 dimer constitutes an integral part of the lipoyl-binding pocket in the N-terminal domain of the opposing subunit. The two swapped C-terminal tails promote conformational changes in active-site clefts of both PDK3 subunits, resulting in largely disordered ATP lids in the ADP-bound form. Our structural and biochemical data suggest that L2 binding stimulates PDK3 activity by disrupting the ATP lid, which otherwise traps ADP, to remove product inhibition exerted by this nucleotide. We hypothesize that this allosteric mechanism accounts, in part, for E2p-augmented PDK3 activity.
- Department of Internal Medicine, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, Dallas, TX 75390, USA.
Organizational Affiliation: