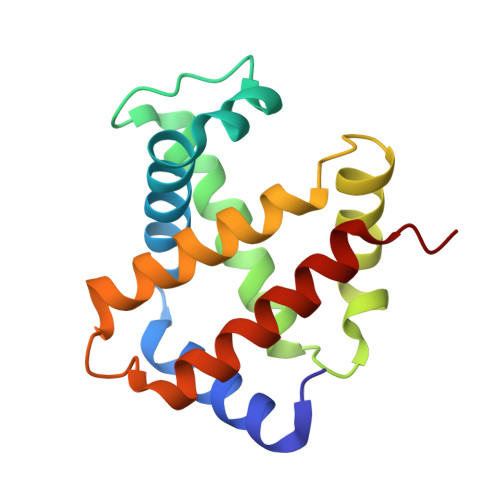
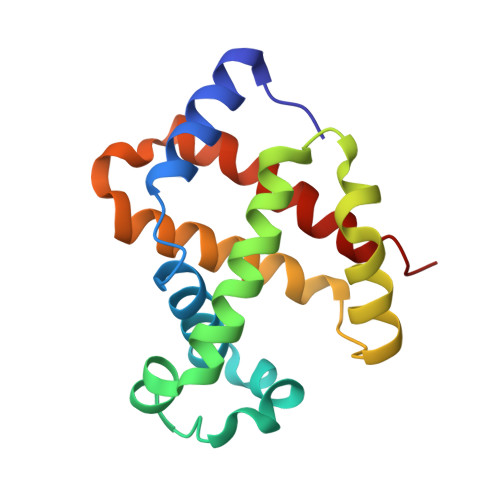
A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Sankaranarayanan, R., Biswal, B.K., Vijayan, M.(2005) Proteins 60: 547-551
- PubMed: 15887226
- DOI: https://doi.org/10.1002/prot.20510
- Primary Citation of Related Structures:
1Y8H, 1Y8I, 1Y8K - PubMed Abstract:
A new relaxed state has been characterized in the crystals of horse methemoglobin grown at neutral pH at low ionic concentration and their low humidity variants. The crystals provide an example for improvement in X-ray diffraction quality with reduced solvent content. Only the classical R state has been so far observed in liganded horse hemoglobin. The state characterized in the present study lies in between the R state and the R2 state characterized earlier in liganded human hemoglobin. The results presented here, along with those of earlier studies, suggest that relaxed and tense hemoglobin can access ensembles of states.
- Molecular Biophysics Unit, Indian Institute of Science, Bangalore, India.
Organizational Affiliation: