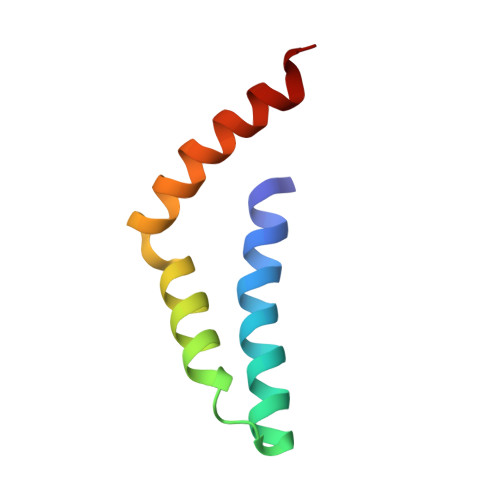
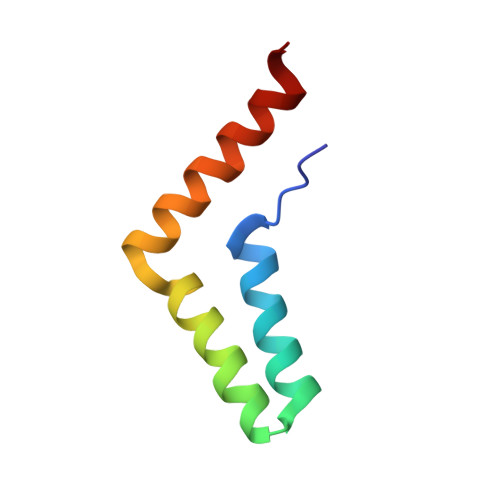
A unified assembly mode revealed by the structures of tetrameric L27 domain complexes formed by mLin-2/mLin-7 and Patj/Pals1 scaffold proteins.
Feng, W., Long, J.F., Zhang, M.(2005) Proc Natl Acad Sci U S A 102: 6861-6866
- PubMed: 15863617
- DOI: https://doi.org/10.1073/pnas.0409346102
- Primary Citation of Related Structures:
1Y74, 1Y76 - PubMed Abstract:
Initially identified in Caenorhabditis elegans Lin-2 and Lin-7, L27 domain is a protein-protein interaction domain capable of organizing scaffold proteins into supramolecular assemblies by formation of heteromeric L27 domain complexes. L27 domain-mediated protein assemblies have been shown to play essential roles in cellular processes including asymmetric cell division, establishment and maintenance of cell polarity, and clustering of receptors and ion channels. The structural basis of L27 domain heteromeric complex assembly is controversial. We determined the high-resolution solution structure of the prototype L27 domain complex formed by mLin-2 and mLin-7 as well as the solution structure of the L27 domain complex formed by Patj and Pals1. The structures suggest that a tetrameric structure composed of two units of heterodimer is a general assembly mode for cognate pairs of L27 domains. Structural analysis of the L27 domain complex structures further showed that the central four-helix bundles mediating tetramer assembly are highly distinct between different pairs of L27 domain complexes. Biochemical studies revealed that the C-terminal alpha-helix responsible for the formation of the central helix bundle is a critical specificity determinant for each L27 domain in choosing its binding partner. Our results provide a unified picture for L27 domain-mediated protein-protein interactions.
- Department of Biochemistry, Molecular Neuroscience Center, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong.
Organizational Affiliation: