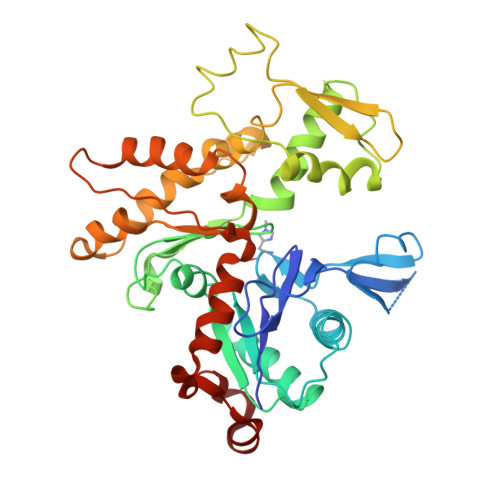
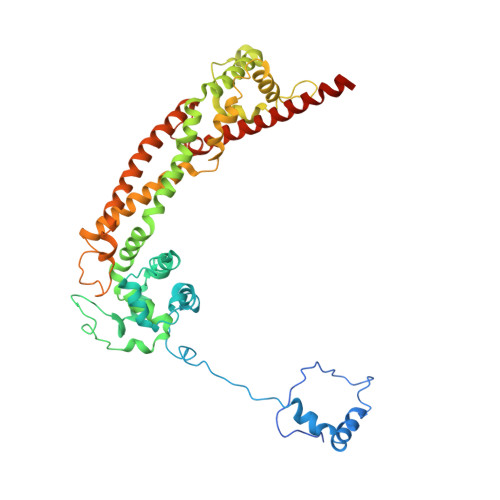
Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain
Otomo, T., Tomchick, D.R., Otomo, C., Panchal, S.C., Machius, M., Rosen, M.K.(2005) Nature 433: 488-494
- PubMed: 15635372
- DOI: https://doi.org/10.1038/nature03251
- Primary Citation of Related Structures:
1Y64 - PubMed Abstract:
The conserved formin homology 2 (FH2) domain nucleates actin filaments and remains bound to the barbed end of the growing filament. Here we report the crystal structure of the yeast Bni1p FH2 domain in complex with tetramethylrhodamine-actin. Each of the two structural units in the FH2 dimer binds two actins in an orientation similar to that in an actin filament, suggesting that this structure could function as a filament nucleus. Biochemical properties of heterodimeric FH2 mutants suggest that the wild-type protein equilibrates between two bound states at the barbed end: one permitting monomer binding and the other permitting monomer dissociation. Interconversion between these states allows processive barbed-end polymerization and depolymerization in the presence of bound FH2 domain. Kinetic and/or thermodynamic differences in the conformational and binding equilibria can explain the variable activity of different FH2 domains as well as the effects of the actin-binding protein profilin on FH2 function.
- Department of Biochemistry, University of Texas Southwestern Medical Center at Dallas, 5323 Harry Hines Boulevard, Dallas, Texas 75390, USA.
Organizational Affiliation: