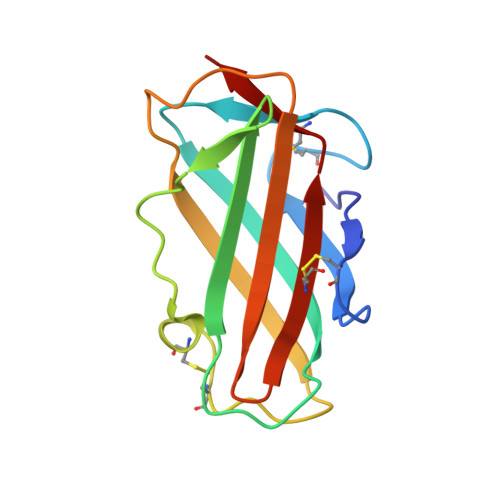
Structure of the house dust mite allergen Der f 2: implications for function and molecular basis of IgE cross-reactivity.
Johannessen, B.R., Skov, L.K., Kastrup, J.S., Kristensen, O., Bolwig, C., Larsen, J.N., Spangfort, M., Lund, K., Gajhede, M.(2005) FEBS Lett 579: 1208-1212
- PubMed: 15710415
- DOI: https://doi.org/10.1016/j.febslet.2004.11.115
- Primary Citation of Related Structures:
1XWV - PubMed Abstract:
The X-ray structure of the group 2 major allergen from Dermatophagoides farinae (Der f 2) was determined to 1.83 A resolution. The overall Der f 2 structure comprises a single domain of immunoglobulin fold with two anti-parallel beta-sheets. A large hydrophobic cavity is formed in the interior of Der f 2. Structural comparisons to distantly related proteins suggest a role in lipid binding. Immunoglobulin E (IgE) cross-reactivity between group 2 house dust mite major allergens can be explained by conserved surface areas representing IgE binding epitopes.
- Biostructural Research, Department of Medicinal Chemistry, Danish University of Pharmaceutical Sciences, Universitetsparken 2, DK-2100 Copenhagen, Denmark.
Organizational Affiliation: