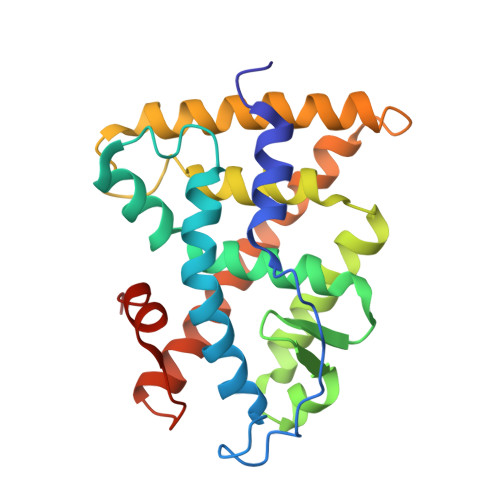
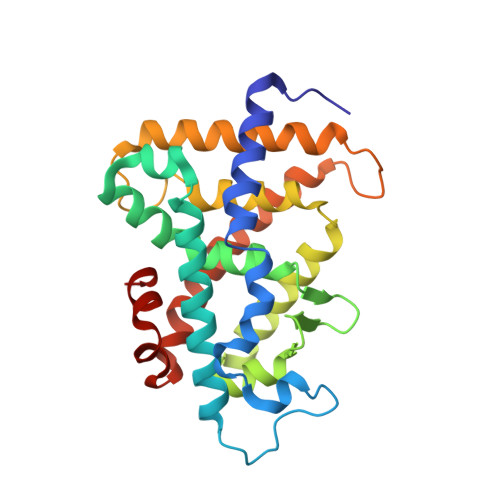
A Structural Basis for Constitutive Activity in the Human CAR/RXRalpha Heterodimer.
Xu, R.X., Lambert, M.H., Wisely, B.B., Warren, E.N., Weinert, E.E., Waitt, G.M., Williams, J.D., Collins, J.L., Moore, L.B., Willson, T.M., Moore, J.T.(2004) Mol Cell 16: 919-928
- PubMed: 15610735
- DOI: https://doi.org/10.1016/j.molcel.2004.11.042
- Primary Citation of Related Structures:
1XV9, 1XVP - PubMed Abstract:
The X-ray crystal structure of the human constitutive androstane receptor (CAR, NR1I3)/retinoid X receptor alpha (RXRalpha, NR2B1) heterodimer sheds light on the mechanism of ligand-independent activation of transcription by nuclear receptors. CAR contains a single-turn Helix X that restricts the conformational freedom of the C-terminal AF2 helix, favoring the active state of the receptor. Helix X and AF2 sit atop four amino acids that shield the CAR ligand binding pocket. A fatty acid ligand was identified in the RXRalpha binding pocket. The endogenous RXRalpha ligand, combined with stabilizing interactions from the heterodimer interface, served to hold RXRalpha in an active conformation. The structure suggests that upon translocation, CAR/RXRalpha heterodimers are preorganized in an active conformation in cells such that they can regulate transcription of target genes. Insights into the molecular basis of CAR constitutive activity can be exploited in the design of inverse agonists as drugs for treatment of obesity.
- Discovery Research, GlaxoSmithKline, Research Triangle Park, NC 27709, USA. robert.x.xu@gsk.com
Organizational Affiliation: