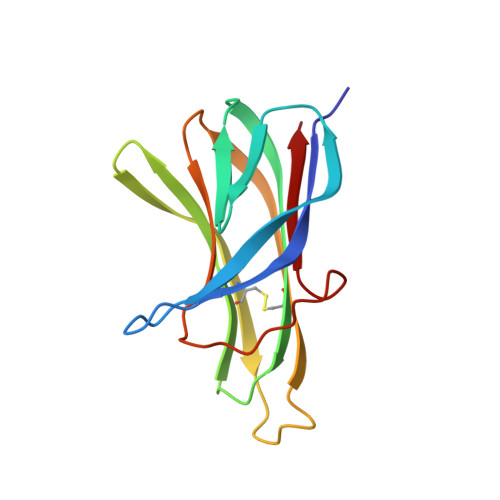
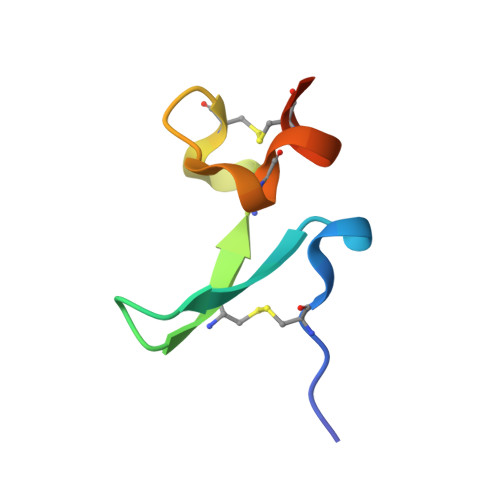
Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
Hymowitz, S.G., Patel, D.R., Wallweber, H.J.A., Runyon, S., Yan, M., Yin, J., Shriver, S.K., Gordon, N.C., Pan, B., Skelton, N.J., Kelley, R.F., Starovasnik, M.A.(2005) J Biological Chem 280: 7218-7227
- PubMed: 15542592
- DOI: https://doi.org/10.1074/jbc.M411714200
- Primary Citation of Related Structures:
1XU1, 1XU2, 1XUT - PubMed Abstract:
TACI is a member of the tumor necrosis factor receptor superfamily and serves as a key regulator of B cell function. TACI binds two ligands, APRIL and BAFF, with high affinity and contains two cysteine-rich domains (CRDs) in its extracellular region; in contrast, BCMA and BR3, the other known high affinity receptors for APRIL and BAFF, respectively, contain only a single or partial CRD. However, another form of TACI exists wherein the N-terminal CRD is removed by alternative splicing. We find that this shorter form is capable of ligand-induced cell signaling and that the second CRD alone (TACI_d2) contains full affinity for both ligands. Furthermore, we report the solution structure and alanine-scanning mutagenesis of TACI_d2 along with co-crystal structures of APRIL.TACI_d2 and APRIL.BCMA complexes that together reveal the mechanism by which TACI engages high affinity ligand binding through a single CRD, and we highlight sources of ligand-receptor specificity within the APRIL/BAFF system.
- Department of Protein Engineering, Molecular Oncology, Medicinal Chemistry, and Immunology, Genentech, Inc., South San Francisco, California 94080, USA.
Organizational Affiliation: