A DNA glycosylase from Pyrobaculum aerophilum with an 8-oxoguanine binding mode and a noncanonical helix-hairpin-helix structure
Lingaraju, G.M., Sartori, A.A., Kostrewa, D., Prota, A.E., Jiricny, J., Winkler, F.K.(2005) Structure 13: 87-98
- PubMed: 15642264
- DOI: https://doi.org/10.1016/j.str.2004.10.011
- Primary Citation of Related Structures:
1XQO, 1XQP - PubMed Abstract:
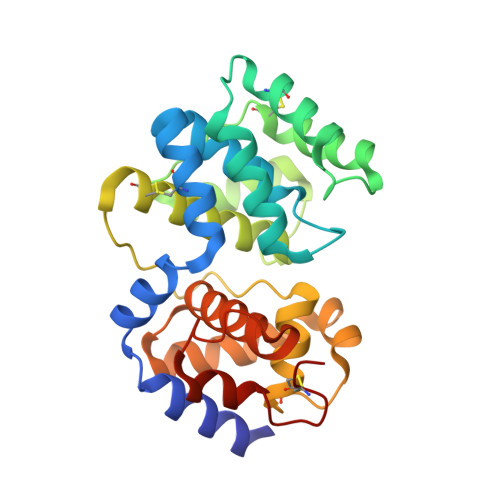
Studies of DNA base excision repair (BER) pathways in the hyperthermophilic crenarchaeon Pyrobaculum aerophilum identified an 8-oxoguanine-DNA glycosylase, Pa-AGOG (archaeal GO glycosylase), with distinct functional characteristics. Here, we describe its crystal structure and that of its complex with 8-oxoguanosine at 1.0 and 1.7 A resolution, respectively. Characteristic structural features are identified that confirm Pa-AGOG to be the founding member of a functional class within the helix-hairpin-helix (HhH) superfamily of DNA repair enzymes. Its hairpin structure differs substantially from that of other proteins containing an HhH motif, and we predict that it interacts with the DNA backbone in a distinct manner. Furthermore, the mode of 8-oxoguanine recognition, which involves several hydrogen-bonding and pi-stacking interactions, is unlike that observed in human OGG1, the prototypic 8-oxoguanine HhH-type DNA glycosylase. Despite these differences, the predicted kinked conformation of bound DNA and the catalytic mechanism are likely to resemble those of human OGG1.
- Biomolecular Research, Paul Scherrer Institut, CH-5232 Villigen, Switzerland.
Organizational Affiliation: