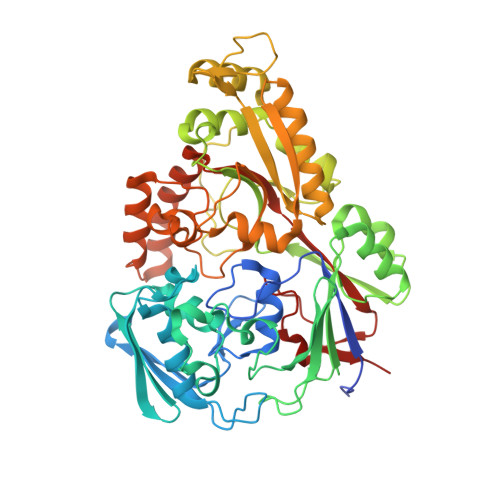
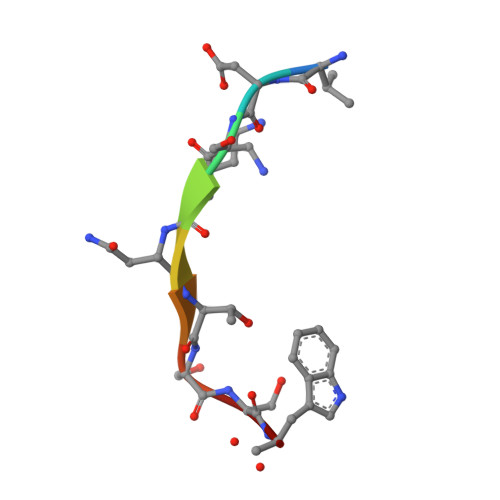
The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide.
Levdikov, V.M., Blagova, E.V., Brannigan, J.A., Wright, L., Vagin, A.A., Wilkinson, A.J.(2005) J Mol Biology 345: 879-892
- PubMed: 15588833
- DOI: https://doi.org/10.1016/j.jmb.2004.10.089
- Primary Citation of Related Structures:
1XOC - PubMed Abstract:
Besides their role as a source of amino acids for Bacillus subtilis, exogenous peptides play important roles in the signalling pathways leading to the development of competence and sporulation. B.subtilis has three peptide transport systems all belonging to the ATP-binding cassette family, a dipeptide permease (Dpp) and two oligopeptide permeases (Opp and App) with overlapping specificity. These comprise a membrane-spanning channel through which the peptide passes, a pair of ATPases which couple ATP hydrolysis to peptide translocation and a lipid-modified, membrane-anchored extracellular "binding-protein" that serves as the receptor for the system. Here, we present the crystal structure of a soluble form of the peptide-binding protein AppA, which has been solved to 1.6 A spacing by anomalous scattering and molecular replacement methods. The structure reveals a protein made of two distinct lobes with a topology similar to those of DppA from Escherichia coli and OppA from Salmonella typhimurium. Examination of the interlobe region reveals an enlarged pocket, containing electron density defining a nonapeptide ligand. The main-chain of the peptide is well defined and makes a series of polar contacts with the protein including salt-bridges at both its termini. The side-chain density is ambiguous in places, consistent with the interpretation that a population of peptides is bound, whose average electron density resembles the amino acid sequence N-VDSKNTSSW-C.
- Structural Biology Laboratory, Department of Chemistry, University of York, York YO10 5YW, UK.
Organizational Affiliation: