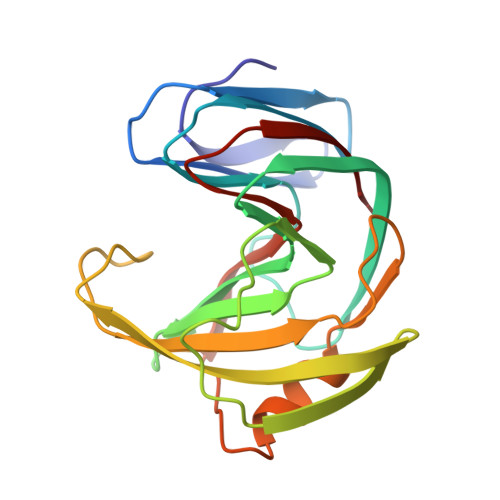
High-Resolution Structures of Xylanases from B. Circulans and T. Harzianum Identify a New Folding Pattern and Implications for the Atomic Basis of the Catalysis
Campbell, R.L., Rose, D.R., Wakarchuk, W.W., To, R.J., Sung, W., Yaguchi, M.To be published.