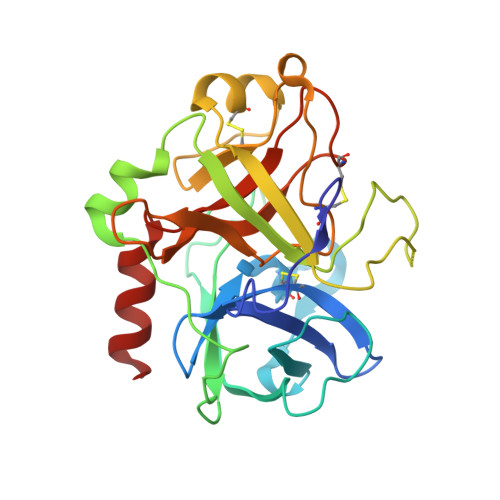
Crystal structure of thrombin bound to heparin
Carter, W.J., Cama, E., Huntington, J.A.(2005) J Biological Chem 280: 2745-2749
- PubMed: 15548541
- DOI: https://doi.org/10.1074/jbc.M411606200
- Primary Citation of Related Structures:
1XMN - PubMed Abstract:
Thrombin is the final protease in the blood coagulation cascade and serves both pro- and anticoagulant functions through the cleavage of several targets. The ability of thrombin to specifically recognize a wide range of substrates derives from interactions that occur outside of the active site of thrombin. Thrombin possesses two anion binding exosites, which mediate many of its interactions with cofactors and substrates, and although many structures of thrombin have been solved, few such interactions have been described in molecular detail. Glycosaminoglycan binding to exosite II of thrombin plays a major role in switching off the procoagulant functions of thrombin by mediating its irreversible inhibition by circulating serpins and by its binding to the endothelial cell surface receptor thrombomodulin. Here we report the 1.85-A structure of human alpha-thrombin bound to a heparin fragment of eight monosaccharide units in length. The asymmetric unit is composed of two thrombin dimers, each sharing a single heparin octasaccharide chain. The observed interactions are fully consistent with previous mutagenesis studies and illustrate on a molecular level the cofactor interaction that is critical for the restriction of clotting to the site of blood vessel injury.
- University of Cambridge, Department of Haematology, Division of Structural Medicine, Thrombosis Research Unit, Cambridge Institute for Medical Research, Wellcome Trust/MRC Building, Hills Road, Cambridge CB2 2XY, United Kingdom.
Organizational Affiliation: