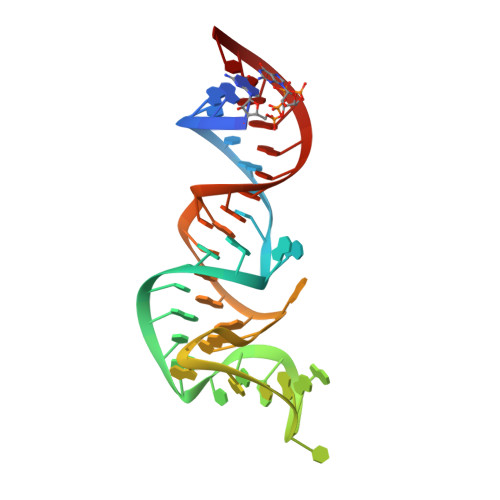
The structure of a rigorously conserved RNA element within the SARS virus genome
Robertson, M.P., Igel, H., Baertsch, R., Haussler, D., Ares Jr., M., Scott, W.G.(2005) PLoS Biol 3: 86-94
- PubMed: 15630477
- DOI: https://doi.org/10.1371/journal.pbio.0030005
- Primary Citation of Related Structures:
1XJR - PubMed Abstract:
We have solved the three-dimensional crystal structure of the stem-loop II motif (s2m) RNA element of the SARS virus genome to 2.7-A resolution. SARS and related coronaviruses and astroviruses all possess a motif at the 3' end of their RNA genomes, called the s2m, whose pathogenic importance is inferred from its rigorous sequence conservation in an otherwise rapidly mutable RNA genome. We find that this extreme conservation is clearly explained by the requirement to form a highly structured RNA whose unique tertiary structure includes a sharp 90 degrees kink of the helix axis and several novel longer-range tertiary interactions. The tertiary base interactions create a tunnel that runs perpendicular to the main helical axis whose interior is negatively charged and binds two magnesium ions. These unusual features likely form interaction surfaces with conserved host cell components or other reactive sites required for virus function. Based on its conservation in viral pathogen genomes and its absence in the human genome, we suggest that these unusual structural features in the s2m RNA element are attractive targets for the design of anti-viral therapeutic agents. Structural genomics has sought to deduce protein function based on three-dimensional homology. Here we have extended this approach to RNA by proposing potential functions for a rigorously conserved set of RNA tertiary structural interactions that occur within the SARS RNA genome itself. Based on tertiary structural comparisons, we propose the s2m RNA binds one or more proteins possessing an oligomer-binding-like fold, and we suggest a possible mechanism for SARS viral RNA hijacking of host protein synthesis, both based upon observed s2m RNA macromolecular mimicry of a relevant ribosomal RNA fold.
- The Center for the Molecular Biology of RNA, University of California, Santa Cruz, USA.
Organizational Affiliation: