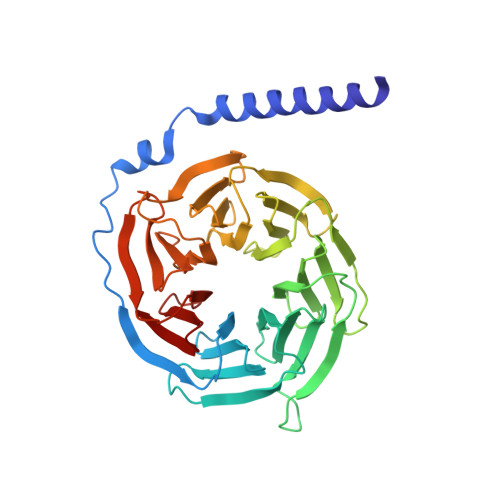
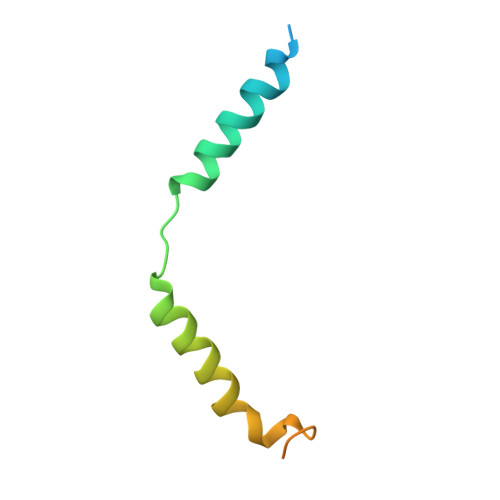
Structural and Molecular Characterization of a Preferred Protein Interaction Surface on G Protein betagamma Subunits.
Davis, T.L., Bonacci, T.M., Sprang, S.R., Smrcka, A.V.(2005) Biochemistry 44: 10593-10604
- PubMed: 16060668
- DOI: https://doi.org/10.1021/bi050655i
- Primary Citation of Related Structures:
1XHM - PubMed Abstract:
G protein betagamma subunits associate with many binding partners in cellular signaling cascades. In previous work, we used random-peptide phage display screening to identify a diverse family of peptides that bound to a common surface on Gbetagamma subunits and blocked a subset of Gbetagamma effectors. Later studies showed that one of the peptides caused G protein activation through a novel Gbetagamma-dependent, nucleotide exchange-independent mechanism. Here we report the X-ray crystal structure of Gbeta(1)gamma(2) bound to this peptide, SIGK (SIGKAFKILGYPDYD), at 2.7 A resolution. SIGK forms a helical structure that binds the same face of Gbeta(1) as the switch II region of Galpha. The interaction interface can be subdivided into polar and nonpolar interfaces that together contain a mixture of binding determinants that may be responsible for the ability of this surface to recognize multiple protein partners. Systematic mutagenic analysis of the peptide-Gbeta(1) interface indicates that distinct sets of amino acids within this interface are required for binding of different peptides. Among these unique amino acid interactions, specific electrostatic binding contacts within the polar interface are required for peptide-mediated subunit dissociation. The data provide a mechanistic basis for multiple target recognition by Gbetagamma subunits with diverse functional interactions within a common interface and suggest that pharmacological targeting of distinct regions within this interface could allow for selective manipulation of Gbetagamma-dependent signaling pathways.
- Department of Biochemistry, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, MC 9050, Dallas, Texas 75390-9050, USA.
Organizational Affiliation: