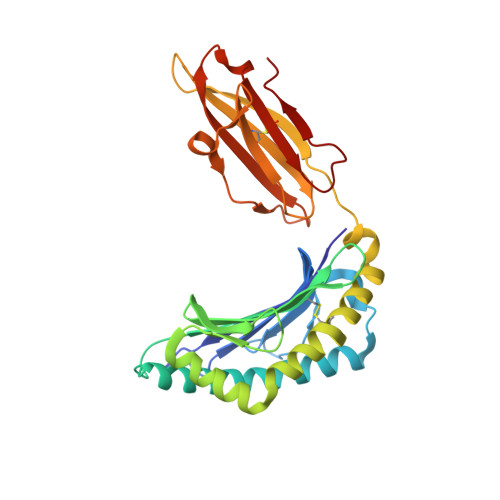
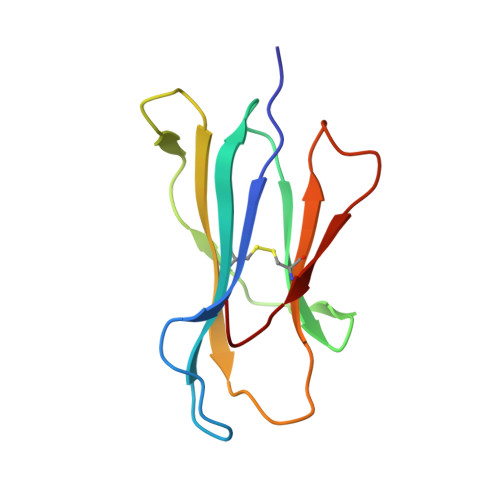
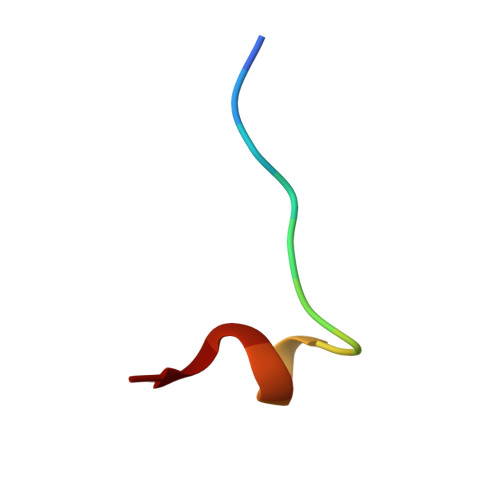
Conformational restraints and flexibility of 14-meric peptides in complex with HLA-B*3501.
Probst-Kepper, M., Hecht, H.J., Herrmann, H., Janke, V., Ocklenburg, F., Klempnauer, J., van den Eynde, B.J., Weiss, S.(2004) J Immunol 173: 5610-5616
- PubMed: 15494511
- DOI: https://doi.org/10.4049/jimmunol.173.9.5610
- Primary Citation of Related Structures:
1XH3 - PubMed Abstract:
Human HLA-B*3501 binds an antigenic peptide of 14-aa length derived from an alternative reading frame of M-CSF with high affinity. Due to its extraordinary length, the exact HLA binding mode was unpredictable. The crystal structure of HLA-B*3501 at 1.5 A shows that the N and C termini of the peptide are embedded in the A and F pockets, respectively, similar to a peptide of normal length. The central part of the 14-meric peptide bulges flexibly out of the groove. Two variants of the alternative reading frame of M-CSF peptide substituted at P2 or P2 and P9 with Ala display weak or no T cell activation. Their structure differs mainly in flexibility and conformation from the agonistic peptide. Moreover, the variants induce subtle changes of MHC alpha-helical regions implicated as critical for TCR contact. The TCR specifically recognizing this peptide/MHC complex exhibits CDR3 length within the normal range, suggesting major conformational adaptations of this receptor upon peptide/MHC binding. Thus, the potential antigenic repertoire recognizable by CTLs is larger than currently thought.
- Department of Visceral and Transplant Surgery, Hannover Medical School, Hannover, Germany. probst-kepper.michael@mh-hannover.de
Organizational Affiliation: