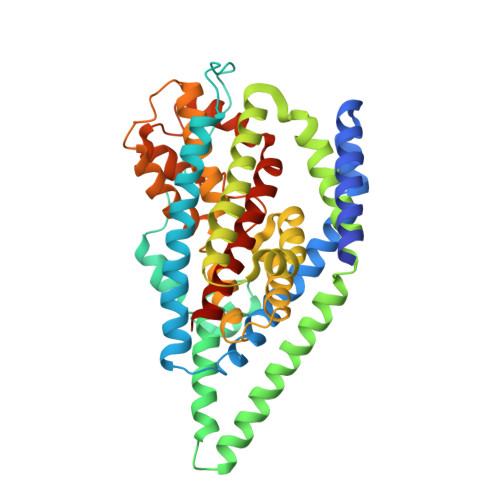
Structure of a glutamate transporter homologue from Pyrococcus horikoshii
Yernool, D., Boudker, O., Jin, Y., Gouaux, E.(2004) Nature 431: 811-818
- PubMed: 15483603
- DOI: https://doi.org/10.1038/nature03018
- Primary Citation of Related Structures:
1XFH - PubMed Abstract:
Glutamate transporters are integral membrane proteins that catalyse the concentrative uptake of glutamate from the synapse to intracellular spaces by harnessing pre-existing ion gradients. In the central nervous system glutamate transporters are essential for normal development and function, and are implicated in stroke, epilepsy and neurodegenerative diseases. Here we present the crystal structure of a eukaryotic glutamate transporter homologue from Pyrococcus horikoshii. The transporter is a bowl-shaped trimer with a solvent-filled extracellular basin extending halfway across the membrane bilayer. At the bottom of the basin are three independent binding sites, each cradled by two helical hairpins, reaching from opposite sides of the membrane. We propose that transport of glutamate is achieved by movements of the hairpins that allow alternating access to either side of the membrane.
- Department of Biochemistry and Molecular Biophysics, Columbia University, 650 West 168th Street, New York, New York 10032, USA.
Organizational Affiliation: