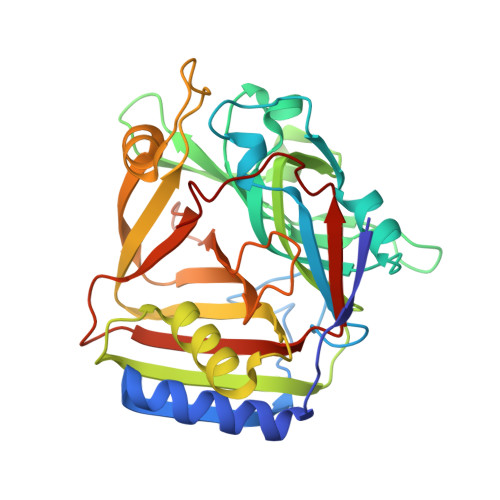
Crystal structure of Homo sapiens PTD012 reveals a zinc-containing hydrolase fold
Manjasetty, B.A., Buessow, K., Fieber-Erdmann, M., Roske, Y., Gobom, J., Scheich, C., Goetz, F., Niesen, F.H., Heinemann, U.(2006) Protein Sci 15: 914-920
- PubMed: 16522806
- DOI: https://doi.org/10.1110/ps.052037006
- Primary Citation of Related Structures:
1XCR - PubMed Abstract:
The human protein PTD012 is the longer product of an alternatively spliced gene and was described to be localized in the nucleus. The X-ray structure analysis at 1.7 A resolution of PTD012 through SAD phasing reveals a monomeric protein and a novel fold. The shorter splice form was also studied and appears to be unfolded and non-functional. The structure of PTD012 displays an alphabetabetaalpha four-layer topology. A metal ion residing between the central beta-sheets is partially coordinated by three histidine residues. X-ray absorption near-edge structure (XANES) analysis identifies the PTD012-bound ion as Zn(2+). Tetrahedral coordination of the ion is completed by the carboxylate oxygen atom of an acetate molecule taken up from the crystallization buffer. The binding of Zn(2+) to PTD012 is reminiscent of zinc-containing enzymes such as carboxypeptidase, carbonic anhydrase, and beta-lactamase. Biochemical assays failed to demonstrate any of these enzyme activities in PTD012. However, PTD012 exhibits ester hydrolase activity on the substrate p-nitrophenyl acetate.
- Protein Structure Factory, Berlin 12489, Germany.
Organizational Affiliation: