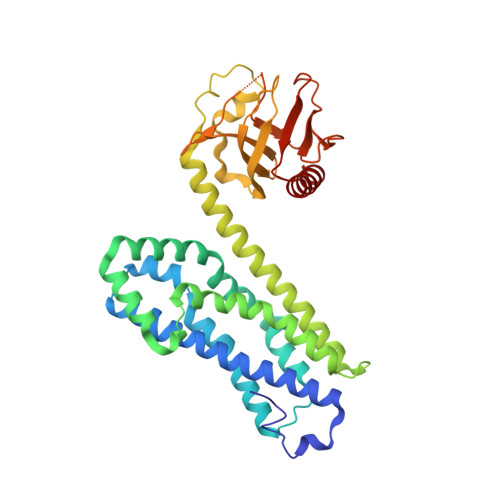
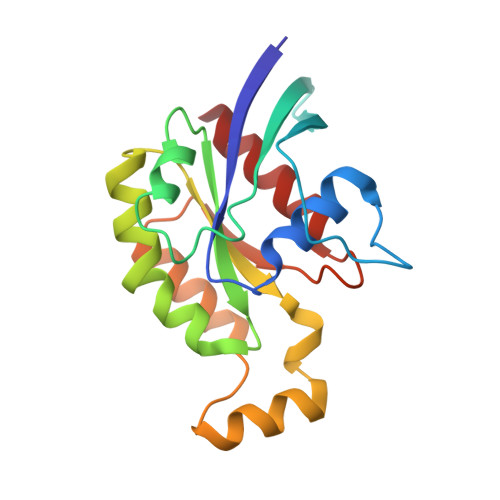
The crystal structure of RhoA in complex with the DH/PH fragment of PDZRhoGEF, an activator of the Ca(2+) sensitization pathway in smooth muscle
Derewenda, U., Oleksy, A., Stevenson, A.S., Korczynska, J., Dauter, Z., Somlyo, A.P., Otlewski, J., Somlyo, A.V., Derewenda, Z.S.(2004) Structure 12: 1955-1965
- PubMed: 15530360
- DOI: https://doi.org/10.1016/j.str.2004.09.003
- Primary Citation of Related Structures:
1XCG - PubMed Abstract:
Calcium sensitization in smooth muscle is mediated by the RhoA GTPase, activated by hitherto unspecified nucleotide exchange factors (GEFs) acting downstream of Galphaq/Galpha(12/13) trimeric G proteins. Here, we show that at least one potential GEF, the PDZRhoGEF, is present in smooth muscle, and its isolated DH/PH fragment induces calcium sensitization in the absence of agonist-mediated signaling. In vitro, the fragment shows high selectivity for the RhoA GTPase. Full-length fragment is required for the nucleotide exchange, as the isolated DH domain enhances it only marginally. We crystallized the DH/PH fragment of PDZRhoGEF in complex with nonprenylated human RhoA and determined the structure at 2.5 A resolution. The refined molecular model reveals that the mutual disposition of the DH and PH domains is significantly different from other previously described complexes involving DH/PH tandems, and that the PH domain interacts with RhoA in a unique mode. The DH domain makes several specific interactions with RhoA residues not conserved among other Rho family members, suggesting the molecular basis for the observed specificity.
- Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, VA 22908, USA.
Organizational Affiliation: