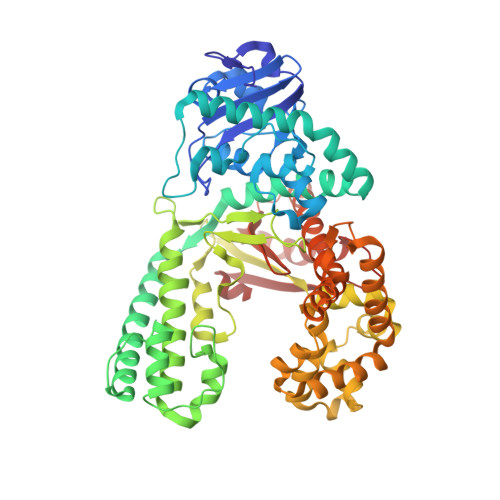
Structure of a High Fidelity DNA Polymerase Bound to a Benzo[a]pyrene Adduct That Blocks Replication
Hsu, G.W., Huang, X., Luneva, N.P., Geacintov, N.E., Beese, L.S.(2005) J Biological Chem 280: 3764-3770
- PubMed: 15548515
- DOI: https://doi.org/10.1074/jbc.M411276200
- Primary Citation of Related Structures:
1XC9 - PubMed Abstract:
Of the carcinogens to which humans are most frequently exposed, the polycyclic aromatic hydrocarbon benzo[a]pyrene (BP) is one of the most ubiquitous. BP is a byproduct of grilled foods and tobacco and fuel combustion and has long been linked to various human cancers, particularly lung and skin. BP is metabolized to diol epoxides that covalently modify DNA bases to form bulky adducts that block DNA synthesis by replicative or high fidelity DNA polymerases. Here we present the structure of a high fidelity polymerase from a thermostable strain of Bacillus stearothermophilus (Bacillus fragment) bound to the most common BP-derived N2-guanine adduct base-paired with cytosine. The BP adduct adopts a conformation that places the polycyclic BP moiety in the nascent DNA minor groove and is the first structure of a minor groove adduct bound to a polymerase. Orientation of the BP moiety into the nascent DNA minor groove results in extensive disruption to the interactions between the adducted DNA duplex and the polymerase. The disruptions revealed by the structure of Bacillus fragment bound to a BP adduct provide a molecular basis for rationalizing the potent blocking effect on replication exerted by BP adducts.
- Department of Biochemistry, Duke University Medical Center, Durham, North Carolina 27710, USA.
Organizational Affiliation: