NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae, and comparison with the X-ray structure of an Aquifex aeolicus homolog
Yeh, D.C., Parsons, L.M., Parsons, J.F., Liu, F., Eisenstein, E., Orban, J.(2005) Protein Sci 14: 424-430
- PubMed: 15632286
- DOI: https://doi.org/10.1110/ps.041096705
- Primary Citation of Related Structures:
1XAX - PubMed Abstract:
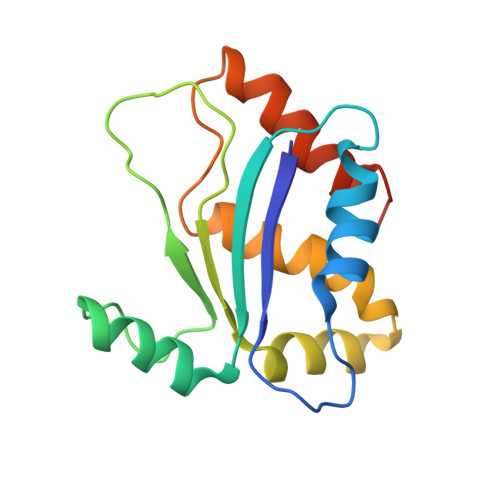
The solution structure of the 154-residue conserved hypothetical protein HI0004 has been determined using multidimensional heteronuclear NMR spectroscopy. HI0004 has sequence homologs in many organisms ranging from bacteria to humans and is believed to be essential in Haemophilus influenzae, although an exact function has yet to be defined. It has a alpha-beta-alpha sandwich architecture consisting of a central four-stranded beta-sheet with the alpha2-helix packed against one side of the beta-sheet and four alpha-helices (alpha1, alpha3, alpha4, alpha5) on the other side. There is structural homology with the eukaryotic matrix metalloproteases (MMPs), but little sequence similarity except for a conserved region containing three histidines that appears in both the MMPs and throughout the HI0004 family of proteins. The solution structure of HI0004 is compared with the X-ray structure of an Aquifex aeolicus homolog, AQ_1354, which has 36% sequence identity over 148 residues. Despite this level of sequence homology, significant differences exist between the two structures. These differences are described along with possible functional implications of the structures.
- Center for Advanced Research in Bio-technology, University of Maryland Biotechnology Institute, 9600 Gudel-sky Drive, Rockville, MD 20850, USA. orban@umbi.umd.edu
Organizational Affiliation: