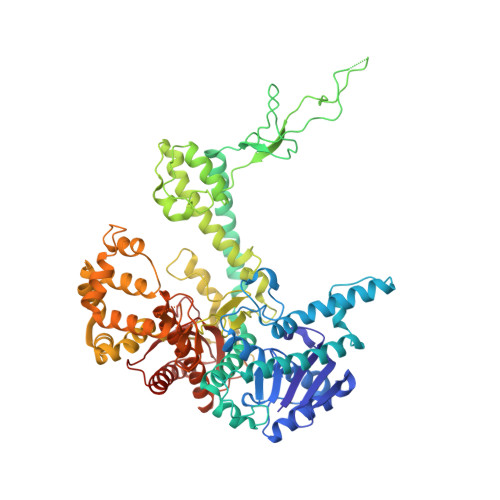
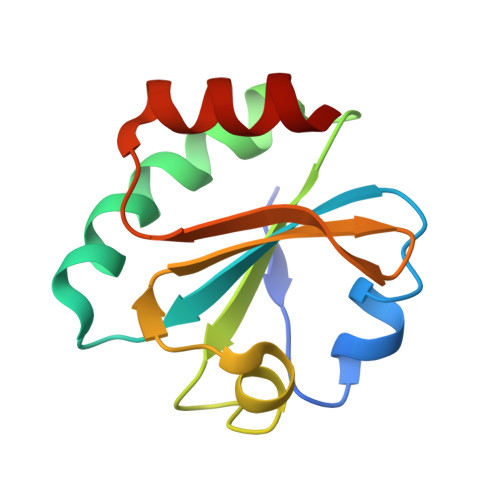
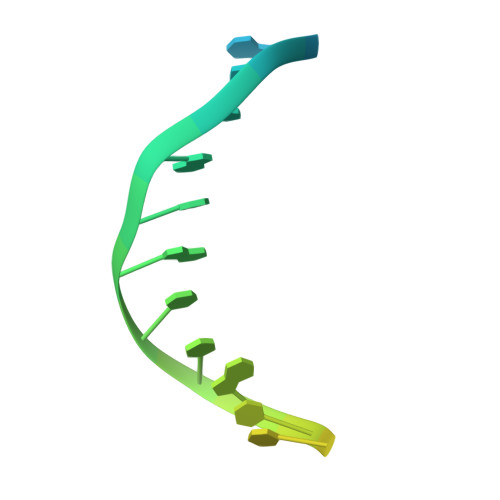
Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis.
Dutta, S., Li, Y., Johnson, D., Dzantiev, L., Richardson, C.C., Romano, L.J., Ellenberger, T.(2004) Proc Natl Acad Sci U S A 101: 16186-16191
- PubMed: 15528277
- DOI: https://doi.org/10.1073/pnas.0406516101
- Primary Citation of Related Structures:
1X9M, 1X9S, 1X9W - PubMed Abstract:
The carcinogen 2-acetylaminofluorene forms two major DNA adducts: N-(2'-deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AAF) and its deacetylated derivative, N-(2'-deoxyguanosin-8-yl)-2-aminofluorene (dG-AF). Although the dG-AAF and dG-AF adducts are distinguished only by the presence or absence of an acetyl group, they have profoundly different effects on DNA replication. dG-AAF poses a strong block to DNA synthesis and primarily induces frameshift mutations in bacteria, resulting in the loss of one or two nucleotides during replication past the lesion. dG-AF is less toxic and more easily bypassed by DNA polymerases, albeit with an increased frequency of misincorporation opposite the lesion, primarily resulting in G --> T transversions. We present three crystal structures of bacteriophage T7 DNA polymerase replication complexes, one with dG-AAF in the templating position and two others with dG-AF in the templating position. Our crystallographic data suggest why a dG-AAF adduct blocks replication more strongly than does a dG-AF adduct and provide a possible explanation for frameshift mutagenesis during replication bypass of a dG-AAF adduct. The dG-AAF nucleoside adopts a syn conformation that facilitates the intercalation of its fluorene ring into a hydrophobic pocket on the surface of the fingers subdomain and locks the fingers in an open, inactive conformation. In contrast, the dG-AF base at the templating position is not well defined by the electron density, consistent with weak binding to the polymerase and a possible interchange of this adduct between the syn and anti conformations.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115, USA.
Organizational Affiliation: