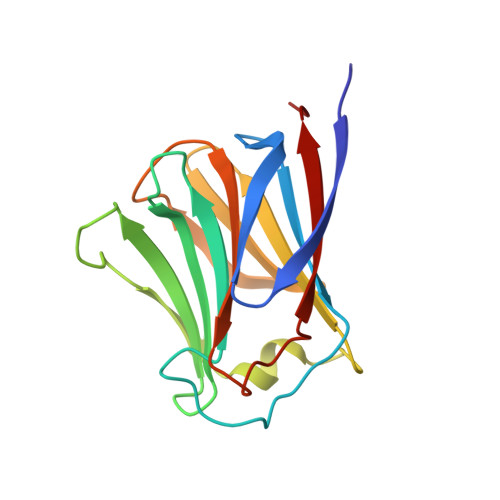
Structural Basis of a Fungal Galectin from Agrocybe cylindracea for Recognizing Sialoconjugate
Ban, M., Yoon, H.J., Demirkan, E., Utsumi, S., Mikami, B., Yagi, F.(2005) J Mol Biology 351: 695-706
- PubMed: 16051274
- DOI: https://doi.org/10.1016/j.jmb.2005.06.045
- Primary Citation of Related Structures:
1WW4, 1WW5, 1WW6, 1WW7 - PubMed Abstract:
Galectin from an edible fungus Agrocybe cylindracea (ACG) has a strong preference for N-acetylneuraminyl lactose (NeuAcalpha2-3lactose). The sugar recognition mechanism of ACG was explored by the X-ray crystallographic analyses of ligand-free ACG, and its complex with lactose, 3'-sulfonyl lactose and NeuAcalpha2-3lactose. The refined structure shows that ACG is a "proto"-type galectin composed of a beta-sandwich of two antiparallel sheets, each with six strands, in contrast to the five and six strands in animal galectins. ACG dimer in solution was classified as being among the "layer"-type. The carbohydrate recognition domain (CRD) of this galectin is common to those of animal galectins, except for substitution of one residue, Ala64, which corresponds to Asn46 in human galectin 1. A five-residue insertion in ACG at positions 42-46 involving Ser44 and Asn46 modified the architecture of the sugar binding site that contributes sialic acid specificity. Furthermore, it was found that the binding of a sulfate ion near the CRD in the ligand-free form led to a change in the conformation of the loop region caused by main-chain cis/trans transition between Ser44 and Pro45.
- Laboratory of Food Design and Development, Graduate school of Agriculture, Kyoto University, Kyoto, Japan.
Organizational Affiliation: