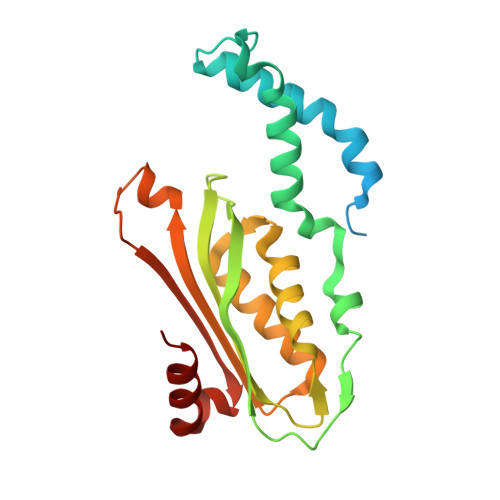
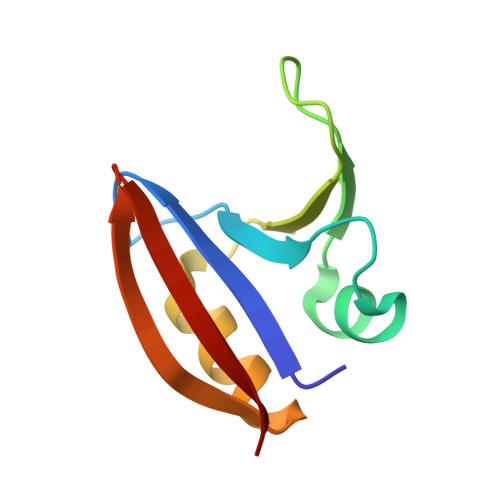
Structural basis of biopterin-induced inhibition of GTP cyclohydrolase I by GFRP, its feedback regulatory protein
Maita, N., Hatakeyama, K., Okada, K., Hakoshima, T.(2004) J Biological Chem 279: 51534-51540
- PubMed: 15448133
- DOI: https://doi.org/10.1074/jbc.M409440200
- Primary Citation of Related Structures:
1WPL - PubMed Abstract:
GTP cyclohydrolase I (GTPCHI) is the rate-limiting enzyme involved in the biosynthesis of tetrahydrobiopterin, a key cofactor necessary for nitric oxide synthase and for the hydroxylases that are involved in the production of catecholamines and serotonin. In animals, the GTPCHI feedback regulatory protein (GFRP) binds GTPCHI to mediate feed-forward activation of GTPCHI activity in the presence of phenylalanine, whereas it induces feedback inhibition of enzyme activity in the presence of biopterin. Here, we have reported the crystal structure of the biopterin-induced inhibitory complex of GTPCHI and GFRP and compared it with the previously reported phenylalanine-induced stimulatory complex. The structure reveals five biopterin molecules located at each interface between GTPCHI and GFRP. Induced fitting structural changes by the biopterin binding expand large conformational changes in GTPCHI peptide segments forming the active site, resulting in inhibition of the activity. By locating 3,4-dihydroxy-phenylalanine-responsive dystonia mutations in the complex structure, we found mutations that may possibly disturb the GFRP-mediated regulation of GTPCHI.
- Structural Biology Laboratory, Nara Institute of Science and Technology, CREST, Japan Science and Technology Agency, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan.
Organizational Affiliation: