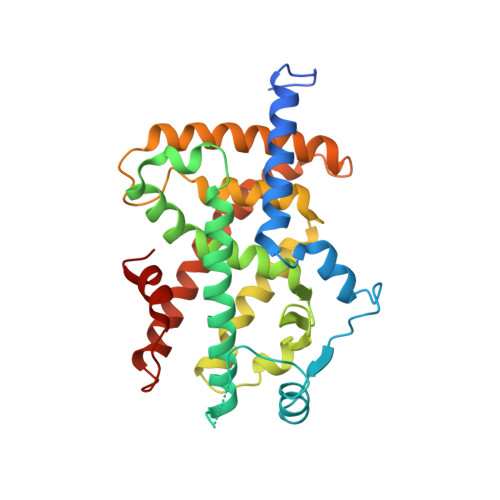
A new class of peroxisome proliferator-activated receptor agonists with a novel binding epitope shows antidiabetic effects
Ostberg, T., Svensson, S., Selen, G., Uppenberg, J., Thor, M., Sundbom, M., Sydow-Backman, M., Gustavsson, A.L., Jendeberg, L.(2004) J Biological Chem 279: 41124-41130
- PubMed: 15258145
- DOI: https://doi.org/10.1074/jbc.M401552200
- Primary Citation of Related Structures:
1WM0 - PubMed Abstract:
The peroxisome proliferator-activated receptors (PPARs) are ligand-activated transcription factors belonging to the NR1 subfamily of nuclear receptors. The PPARs play key roles in the control of glucose and lipid homeostasis, and the synthetic isoform-specific PPAR agonists are used clinically to improve insulin sensitivity and to lower serum triglyceride levels. All of the previously reported PPAR agonists form the same characteristic interactions with the receptor, which have been postulated to be important for the induction of agonistic activity. Here we describe a new class of PPARalpha/gamma modulators, the 5-substituted 2-benzoylaminobenzoic acids (2-BABAs). As shown by x-ray crystallography, the representative compounds BVT.13, BVT.762, and BVT.763, utilize a novel binding epitope and lack the agonist-characteristic interactions. Despite this, some compounds within the 2-BABA family are potent agonists in a cell-based reporter gene assay. Furthermore, BVT.13 displays antidiabetic effects in ob/ob mice. We concluded that the 2-BABA binding mode can be used to design isoform-specific PPAR modulators with biological activity in vivo.
- Department of Cell and Molecular Biology, Medical Nobel Institute, Karolinska Institute, SE-171 77 Stockholm, Sweden.
Organizational Affiliation: