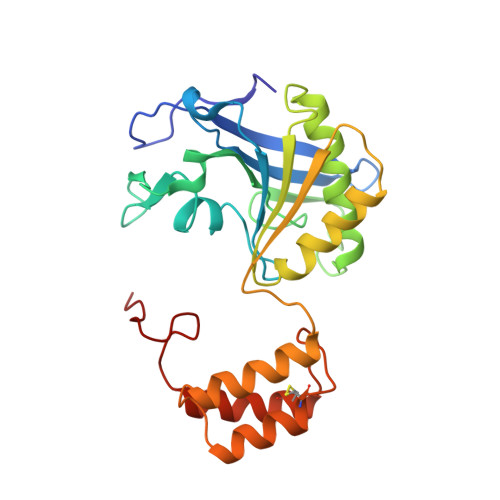
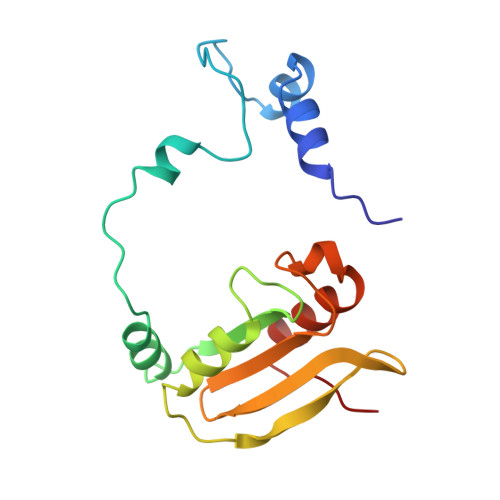
Structure of the complex of L-benzylsuccinate with wheat serine carboxypeptidase II at 2.0-A resolution.
Bullock, T.L., Branchaud, B., Remington, S.J.(1994) Biochemistry 33: 11127-11134
- PubMed: 7727364
- DOI: https://doi.org/10.1021/bi00203a009
- Primary Citation of Related Structures:
1WHS, 1WHT - PubMed Abstract:
The structure of the complex of L-benzylsuccinate (Ki = 0.2 mM) bound to wheat serine carboxypeptidase II has been analyzed at 2.0-A resolution for native and inhibited crystals at -170 degrees C. The model has been refined and has a standard crystallographic R-factor of 0.176 for 57,734 reflections observed between 20.0- and 2.0-A resolution. The root mean square deviation from ideal bonds is 0.017 A and from ideal angles is 2.6 degrees. The model consists of 400 amino acids, 4 N-linked saccharide residues, and 430 water molecules. L-Benzylsuccinate occupies a narrow slot in the active site defined by Tyr 60, Tyr 239, and the polypeptide backbone. One carboxylate forms hydrogen bonds to Glu 145, Asn 51, the amide of Gly 52, and the catalytic His 397, suggestive of how the peptide C-terminal carboxylate is recognized by the enzyme. The phenyl ring stacks between Tyr 239 and Tyr 60, while the other carboxylate occupies the "oxyanion hole". One of the oxygens accepts hydrogen bonds from the amides of Tyr 147 and Gly 53, while the other forms a very close contact (2.3 A) with the O gamma of Ser 146, forcing the side chain into a conformation alternative to that found in the resting state of the enzyme. The inhibitor occupies the active site in a way that suggests that it can be regarded as a transition-state analogue of serine carboxypeptidases. The model suggests a novel enzymatic mechanism, involving substrate-assisted catalysis, that might account for the low pH optimum (4.0-5.5) of peptidase activity unique to this family of serine proteinases.
- Institute of Molecular Biology, University of Oregon, Eugene 97403, USA.
Organizational Affiliation: