Crystal Structure of the Mosquito-Larvicidal Toxin Cry4Ba and its Biological Implications
Boonserm, P., Davis, P., Ellar, D.J., Li, J.(2005) J Mol Biology 348: 363
- PubMed: 15811374
- DOI: https://doi.org/10.1016/j.jmb.2005.02.013
- Primary Citation of Related Structures:
1W99 - PubMed Abstract:
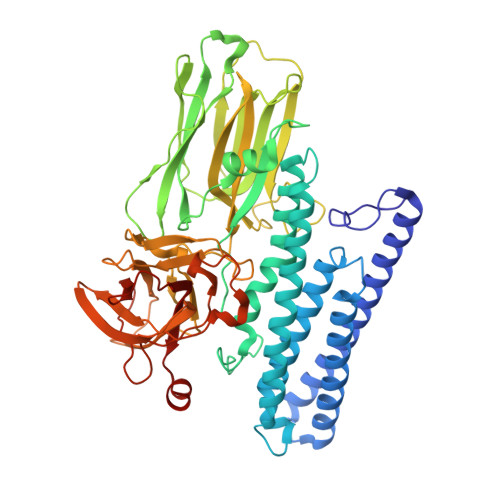
Cry4Ba, isolated from Bacillus thuringiensis subsp. israelensis, is specifically toxic to the larvae of Aedes and Anopheles mosquitoes. The structure of activated Cry4Ba toxin has been determined by multiple isomorphous replacement with anomalous scattering and refined to R(cryst) = 20.5% and R(free)= 21.8% at 1.75 Angstroms resolution. It resembles previously reported Cry toxin structures but shows the following distinctions. In domain I the helix bundle contains only the long and amphipathic helices alpha3-alpha7. The N-terminal helices alpha1-alpha2b, absent due to proteolysis during crystallisation, appear inessential to toxicity. In domain II the beta-sheet prism presents short apical loops without the beta-ribbon extension of inner strands, thus placing the receptor combining sites close to the sheets. In domain III the beta-sandwich contains a helical extension from the C-terminal strand beta23, which interacts with a beta-hairpin excursion from the edge of the outer sheet. The structure provides a rational explanation of recent mutagenesis and biophysical data on this toxin. Furthermore, added to earlier structures from the Cry toxin family, Cry4Ba completes a minimal structural database covering the Coleoptera, Lepidoptera, Diptera and Lepidoptera/Diptera specificity classes. A multiple structure alignment found that the Diptera-specific Cry4Ba is structurally more closely similar to the Lepidoptera-specific Cry1Aa than the Coleoptera-specific Cry3Aa, but most distantly related to Lepidoptera/Diptera-specific Cry2Aa. The structures are most divergent in domain II, supporting the suggestion that this domain has a major role in specificity determination. They are most similar in the alpha3-alpha7 major fragment of domain I, which contains the alpha4-alpha5 hairpin crucial to pore formation. The collective knowledge of Cry toxin structure and mutagenesis data will lead to a more critical understanding of the structural basis for receptor binding and pore formation, as well as allowing the scope of diversity to be better appreciated.
- Department of Biochemistry, University of Cambridge, Cambridge CB2 1GA, UK.
Organizational Affiliation: