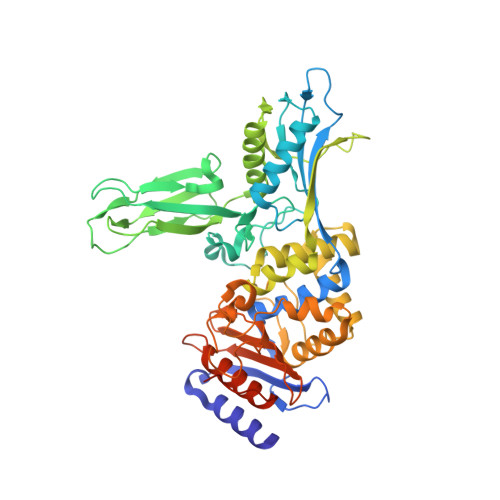
Crystal Structure of the Actinomadura R39 Dd-Peptidase Reveals New Domains in Penicillin-Binding Proteins.
Sauvage, E., Herman, R., Petrella, S., Duez, C., Bouillenne, F., Frere, J.M., Charlier, P.(2005) J Biological Chem 280: 31249
- PubMed: 15987687
- DOI: https://doi.org/10.1074/jbc.M503271200
- Primary Citation of Related Structures:
1W79, 1W8Q, 1W8Y - PubMed Abstract:
Actinomadura sp. R39 produces an exocellular DD-peptidase/penicillin-binding protein (PBP) whose primary structure is similar to that of Escherichia coli PBP4. It is characterized by a high beta-lactam-binding activity (second order rate constant for the acylation of the active site serine by benzylpenicillin: k2/K = 300 mm(-1) s(-1)). The crystal structure of the DD-peptidase from Actinomadura R39 was solved at a resolution of 1.8 angstroms by single anomalous dispersion at the cobalt resonance wavelength. The structure is composed of three domains: a penicillin-binding domain similar to the penicillin-binding domain of E. coli PBP5 and two domains of unknown function. In most multimodular PBPs, additional domains are generally located at the C or N termini of the penicillin-binding domain. In R39, the other two domains are inserted in the penicillin-binding domain, between the SXXK and SXN motifs, in a manner similar to "Matryoshka dolls." One of these domains is composed of a five-stranded beta-sheet with two helices on one side, and the other domain is a double three-stranded beta-sheet inserted in the previous domain. Additionally, the 2.4-angstroms structure of the acyl-enzyme complex of R39 with nitrocefin reveals the absence of active site conformational change upon binding the beta-lactams.
- Centre d'Ingénierie des Protéines, Université de Liège, Institut de Physique B5, B-4000 Liège, Belgium. eric.sauvage@ulg.ac.be
Organizational Affiliation: