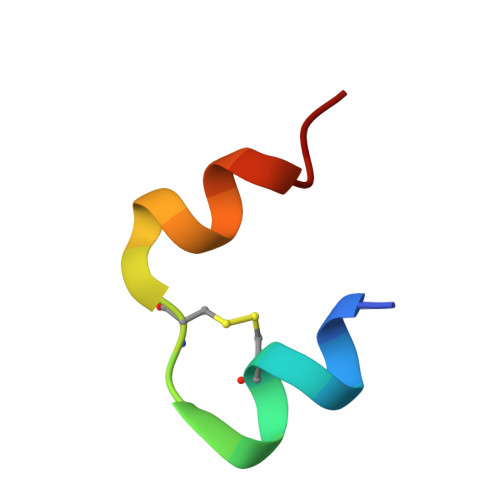
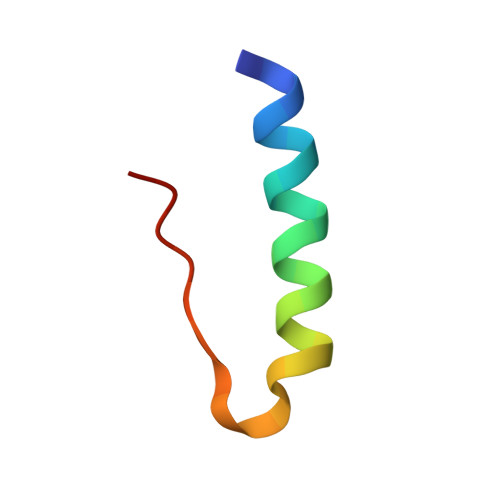
Towards the Insulin-Igf-I Intermediate Structures: Functional and Structural Properties of the B25Tyr-Nme-B26Phe Insulin Mutant.
Zarkowa, L., Brynda, J., Au-Alvarez, O., Dodson, E.J., Dodson, G.G., Whittingham, J.L., Brzozowski, A.M.(2004) Biochemistry 43: 16293
- PubMed: 15610023
- DOI: https://doi.org/10.1021/bi048856u
- Primary Citation of Related Structures:
1W8P - PubMed Abstract:
The origins of differentiation of insulin from insulin-like growth factor I (IGF-I) are still unknown. To address the problem of a structural and biological switch from the mostly metabolic hormonal activity of insulin to the predominant growth factor activities of IGF-I, an insulin analogue with IGF-I-like structural features has been synthesized. Insulin residues Phe(B25) and Tyr(B26) have been swapped with the IGF-I-like Tyr(24) and Phe(25) sequence with a simultaneous methylation of the peptide nitrogen of residue Phe(B26). These modifications were expected to introduce a substantial kink in the main chain, as observed at residue Phe(25) in the IGF-I crystal structure. These alterations should provide insight into the structural origins of insulin-IGF-I structural and functional divergence. The [Tyr(B25)NMePhe(B26)] mutant has been characterized, and its crystal structure has been determined. Surprisingly, all of these changes are well accommodated within an insulin R6 hexamer. Only one molecule of each dimer in the hexamer responds to the structural alterations, the other remaining very similar to wild-type insulin. All alterations, modest in their scale, cumulate in the C-terminal part of the B-chain (residues B23-B30), which moves toward the core of the insulin molecule and is associated with a significant shift of the A1 helix toward the C-terminus of the B-chain. These changes do not produce the expected bend of the main chain, but the fold of the mutant does reflect some structural characteristics of IGF-1, and in addition establishes the CO(A19)-NH(B25) hydrogen bond, which is normally characteristic of T-state insulin.
- Department of Biological Chemistry, Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, Flemingovo 2, 166 10 Prague, Czech Republic.
Organizational Affiliation: