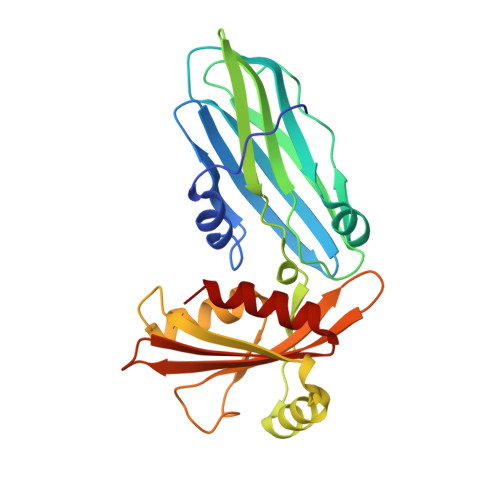
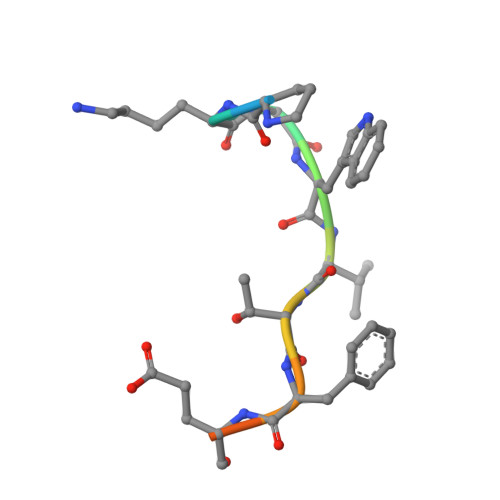
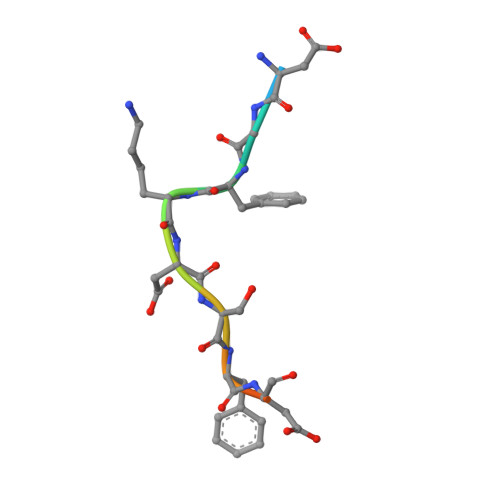
Evolving Nature of the Ap2 Alpha-Appendage Hub During Clathrin-Coated Vesicle Endocytosis.
Praefcke, G.J.K., Ford, M.G.J., Schmid, E.M., Olesen, L.E., Gallop, J.L., Peak-Chew, S.-Y., Vallis, Y., Babu, M.M., Mills, I.G., Mcmahon, H.T.(2004) EMBO J 23: 4371
- PubMed: 15496985
- DOI: https://doi.org/10.1038/sj.emboj.7600445
- Primary Citation of Related Structures:
1W80 - PubMed Abstract:
Clathrin-mediated endocytosis involves the assembly of a network of proteins that select cargo, modify membrane shape and drive invagination, vesicle scission and uncoating. This network is initially assembled around adaptor protein (AP) appendage domains, which are protein interaction hubs. Using crystallography, we show that FxDxF and WVxF peptide motifs from synaptojanin bind to distinct subdomains on alpha-appendages, called 'top' and 'side' sites. Appendages use both these sites to interact with their binding partners in vitro and in vivo. Occupation of both sites simultaneously results in high-affinity reversible interactions with lone appendages (e.g. eps15 and epsin1). Proteins with multiple copies of only one type of motif bind multiple appendages and so will aid adaptor clustering. These clustered alpha(appendage)-hubs have altered properties where they can sample many different binding partners, which in turn can interact with each other and indirectly with clathrin. In the final coated vesicle, most appendage binding partners are absent and thus the functional status of the appendage domain as an interaction hub is temporal and transitory giving directionality to vesicle assembly.
- Medical Research Council Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: