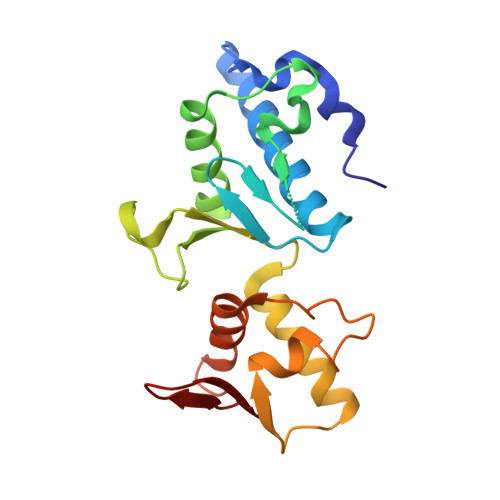
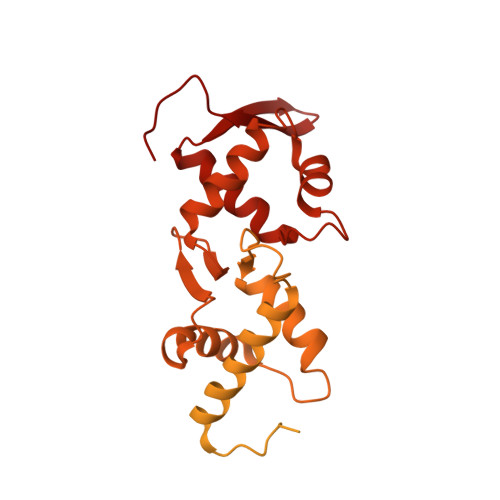
Escrt-II, an Endosome-Associated Complex Required for Protein Sorting: Crystal Structure and Interactions with Escrt-III and Membranes
Teo, H., Perisic, O., Gonzalez, B., Williams, R.L.(2004) Dev Cell 7: 559
- PubMed: 15469844
- DOI: https://doi.org/10.1016/j.devcel.2004.09.003
- Primary Citation of Related Structures:
1W7P - PubMed Abstract:
ESCRT-I, -II, and -III protein complexes are sequentially recruited to endosomal membranes, where they orchestrate protein sorting and MVB biogenesis. In addition, they play a critical role in retrovirus budding. Structural understanding of ESCRT interaction networks is largely lacking. The 3.6 A structure of the yeast ESCRT-II core presented here reveals a trilobal complex containing two copies of Vps25, one copy of Vps22, and the C-terminal region of Vps36. Unexpectedly, the entire ESCRT-II core consists of eight repeats of a common building block, a "winged helix" domain. Two PPXY-motifs from Vps25 are involved in contacts with Vps22 and Vps36, and their mutation leads to ESCRT-II disruption. We show that purified ESCRT-II binds directly to the Vps20 component of ESCRT-III. Surprisingly, this binding does not require the protruding N-terminal coiled-coil of Vps22. Vps25 is the chief subunit responsible for Vps20 recruitment. This interaction dramatically increases binding of both components to lipid vesicles in vitro.
- MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, United Kingdom.
Organizational Affiliation: