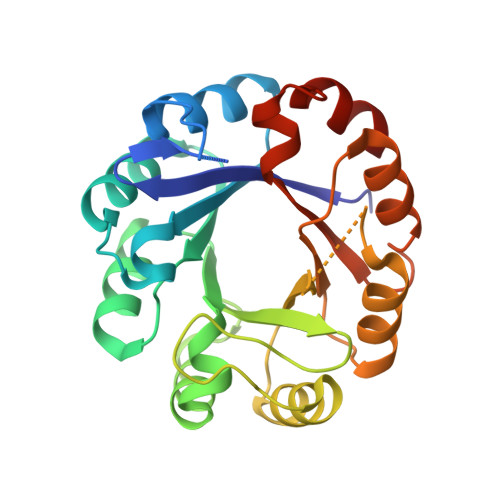
Two-Fold Repeated (Beta-Alpha)(4) Half-Barrels May Provide a Molecular Tool for Dual Substrate Specificity
Kuper, J., Doenges, C., Wilmanns, M.(2005) EMBO Rep 6: 134
- PubMed: 15654319
- DOI: https://doi.org/10.1038/sj.embor.7400330
- Primary Citation of Related Structures:
1VZW - PubMed Abstract:
Some bacterial genomes contain an incomplete set of genes encoding phosphoribosyl isomerases, raising the question of whether there exists broadened substrate specificity for the missing gene products. To investigate the underlying molecular principles of this hypothesis, we have determined the crystal structure of the bifunctional enzyme PriA from Streptomyces coelicolor at 1.8 A resolution. It consists of a (betaalpha)(8)-barrel fold that is assembled by two symmetric (betaalpha)(4) half-barrels. The structure shows how its active site may catalyse the isomerization reactions of two different substrates, and we provide a plausible model of how the smaller of the two substrates could be bound in two different orientations. Our findings expand the half-barrel ancestor concept by demonstrating that symmetry-related half-barrels could provide a smart solution to cope with dual substrate specificity. The data may help to unravel molecular rationales regarding how organisms with miniature genomes can keep central biological pathways functional.
- EMBL-Hamburg Outstation, DESY, Notkestrasse 85, 22603 Hamburg, Germany.
Organizational Affiliation: