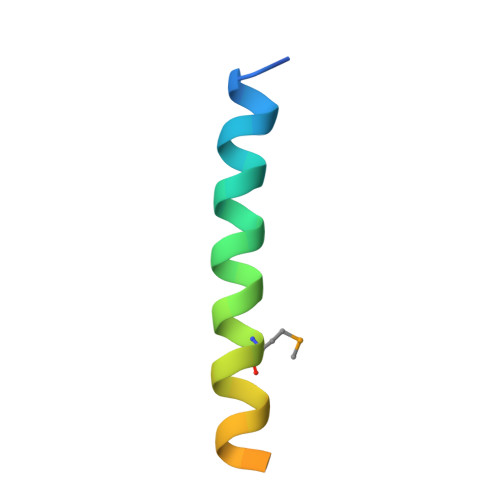
The Synaptic Acetylcholinesterase Tetramer Assembles Around a Polyproline-II Helix
Dvir, H., Harel, M., Bon, S., Liu, W.-Q., Vidal, M., Garbay, C., Sussman, J.L., Massoulie, J., Silman, I.(2004) EMBO J 23: 4394
- PubMed: 15526038
- DOI: https://doi.org/10.1038/sj.emboj.7600425
- Primary Citation of Related Structures:
1VZJ - PubMed Abstract:
Functional localization of acetylcholinesterase (AChE) in vertebrate muscle and brain depends on interaction of the tryptophan amphiphilic tetramerization (WAT) sequence, at the C-terminus of its major splice variant (T), with a proline-rich attachment domain (PRAD), of the anchoring proteins, collagenous (ColQ) and proline-rich membrane anchor. The crystal structure of the WAT/PRAD complex reveals a novel supercoil structure in which four parallel WAT chains form a left-handed superhelix around an antiparallel left-handed PRAD helix resembling polyproline II. The WAT coiled coils possess a WWW motif making repetitive hydrophobic stacking and hydrogen-bond interactions with the PRAD. The WAT chains are related by an approximately 4-fold screw axis around the PRAD. Each WAT makes similar but unique interactions, consistent with an asymmetric pattern of disulfide linkages between the AChE tetramer subunits and ColQ. The P59Q mutation in ColQ, which causes congenital endplate AChE deficiency, and is located within the PRAD, disrupts crucial WAT-WAT and WAT-PRAD interactions. A model is proposed for the synaptic AChE(T) tetramer.
- Dapartment of Structural Biology, Weizmann Institute of Science, Rehovot, Israel.
Organizational Affiliation: