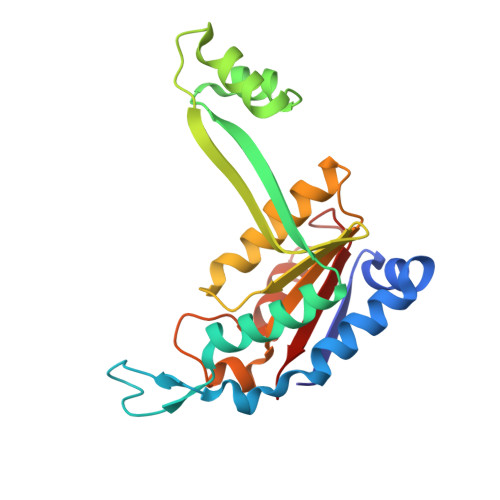
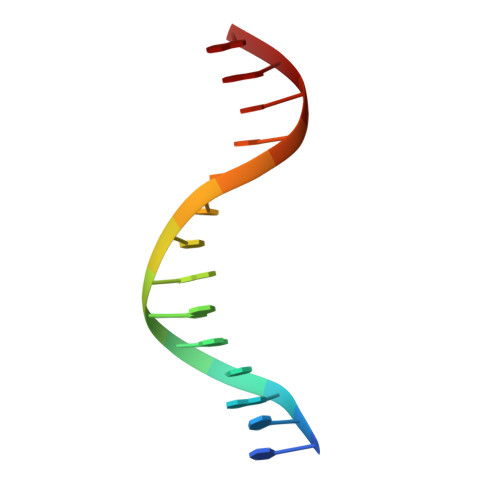
Implications for Switching Restriction Enzyme Specificities from the Structure of BstYI Bound to a BglII DNA Sequence.
Townson, S.A., Samuelson, J.C., Xu, S.Y., Aggarwal, A.K.(2005) Structure 13: 791-801
- PubMed: 15893669
- DOI: https://doi.org/10.1016/j.str.2005.02.018
- Primary Citation of Related Structures:
1VRR - PubMed Abstract:
The type II restriction endonuclease BstYI recognizes the degenerate sequence 5'-RGATCY-3' (where R = A/G and Y = C/T), which overlaps with both BamHI (GGATCC) and BglII (AGATCT), and thus raises the question of whether BstYI DNA recognition will be more BamHI-like or BglII-like. We present here the structure of BstYI bound to a cognate DNA sequence (AGATCT). We find the complex to be more BglII-like with similarities mapping to DNA conformation, domain organization, and residues involved in catalysis. However, BstYI is unique in containing an extended arm subdomain, and the mechanism of DNA capture has both BglII-like and BamHI-like elements. Further, DNA recognition is more minimal than BglII and BamHI, where only two residues mediate recognition of the entire core sequence. Taken together, the structure reveals a mechanism of degenerate DNA recognition and offers insights into the possibilities and limitations in altering specificities of closely related restriction enzymes.
- Structural Biology Program, Department of Physiology & Biophysics, Mount Sinai School of Medicine, 1425 Madison Avenue, New York, New York 10029, USA.
Organizational Affiliation: