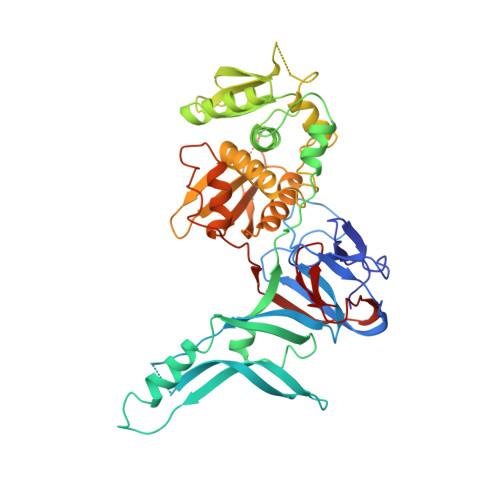
Crystal structure of PI-SceI, a homing endonuclease with protein splicing activity.
Duan, X., Gimble, F.S., Quiocho, F.A.(1997) Cell 89: 555-564
- PubMed: 9160747
- DOI: https://doi.org/10.1016/s0092-8674(00)80237-8
- Primary Citation of Related Structures:
1VDE - PubMed Abstract:
PI-Scel is a bifunctional yeast protein that propagates its mobile gene by catalyzing protein splicing and site-specific DNA double-strand cleavage. Here, we report the 2.4 A crystal structure of the PI-Scel protein. The structure is composed of two separate domains (I and II) with novel folds and different functions. Domain I, which is elongated and formed largely from seven beta sheets, harbors the N and C termini residues and two His residues that are implicated in protein splicing. Domain II, which is compact and is primarily composed of two similar alpha/beta motifs related by local two-fold symmetry, contains the putative nuclease active site with a cluster of two acidic residues and one basic residue commonly found in restriction endonucleases. This report presents prototypic structures of domains with single endonuclease and protein splicing active sites.
- Structural and Computational Biology and Molecular Biophysics Program, Baylor College of Medicine, Houston, Texas 77030, USA.
Organizational Affiliation: