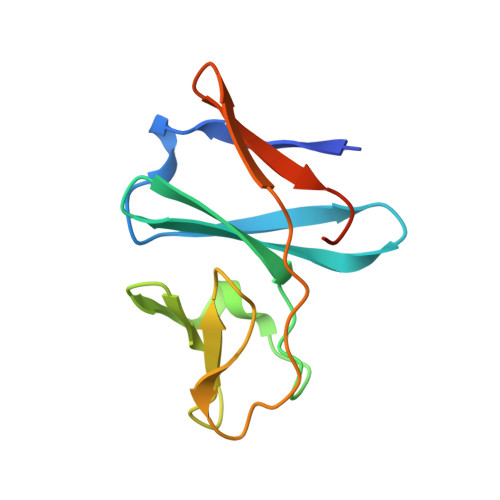
Crystal structure of the ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10, a novel Rieske non-heme iron oxygenase system
Nam, J.-W., Noguchi, H., Fujiomoto, Z., Mizuno, H., Ashikawa, Y., Abo, M., Fushinobu, S., Kobashi, N., Wakagi, T., Iwata, K., Yoshida, T., Habe, H., Yamane, H., Omori, T., Nojiri, H.(2005) Proteins 58: 779-789
- PubMed: 15645447
- DOI: https://doi.org/10.1002/prot.20374
- Primary Citation of Related Structures:
1VCK - PubMed Abstract:
The carbazole 1,9a-dioxygenase (CARDO) system of Pseudomonas resinovorans strain CA10 catalyzes the dioxygenation of carbazole; the 9aC carbon bonds to a nitrogen atom and its adjacent 1C carbon as the initial reaction in the mineralization pathway. The CARDO system is composed of ferredoxin reductase (CarAd), ferredoxin (CarAc), and terminal oxygenase (CarAa). CarAc acts as a mediator in the electron transfer from CarAd to CarAa. To understand the structural basis of the protein-protein interactions during electron transport in the CARDO system, the crystal structure of CarAc was determined at 1.9 A resolution by molecular replacement using the structure of BphF, the biphenyl 2,3-dioxygenase ferredoxin from Burkholderia cepacia strain LB400 as a search model. CarAc is composed of three beta-sheets, and the structure can be divided into two domains, a cluster-binding domain and a basal domain. The Rieske [2Fe-2S] cluster is located at the tip of the cluster-binding domain, where it is exposed to solvent. While the overall folding of CarAc and BphF is strongly conserved, the properties of their surfaces are very different from each other. The structure of the cluster-binding domain of CarAc is more compact and protruding than that of BphF, and the distribution of electric charge on its molecular surface is very different. Such differences are thought to explain why these ferredoxins can act as electron mediators in respective electron transport chains composed of different-featured components.
- Biotechnology Research Center, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo, Japan.
Organizational Affiliation: