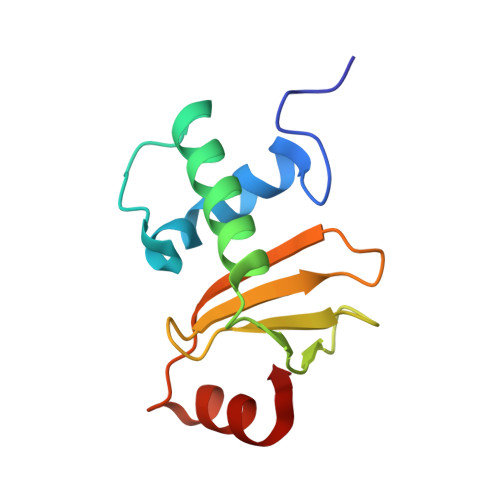
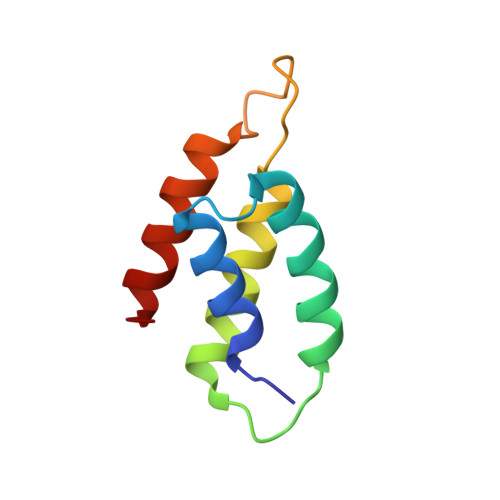
Structural inhibition of the colicin D tRNase by the tRNA-mimicking immunity protein
Graille, M., Mora, L., Buckingham, R.H., Van Tilbeurgh, H., De Zamaroczy, M.(2004) EMBO J 23: 1474-1482
- PubMed: 15014439
- DOI: https://doi.org/10.1038/sj.emboj.7600162
- Primary Citation of Related Structures:
1V74 - PubMed Abstract:
Colicins are toxins secreted by Escherichia coli in order to kill their competitors. Colicin D is a 75 kDa protein that consists of a translocation domain, a receptor-binding domain and a cytotoxic domain, which specifically cleaves the anticodon loop of all four tRNA(Arg) isoacceptors, thereby inactivating protein synthesis and leading to cell death. Here we report the 2.0 A resolution crystal structure of the complex between the toxic domain and its immunity protein ImmD. Neither component shows structural homology to known RNases or their inhibitors. In contrast to other characterized colicin nuclease-Imm complexes, the colicin D active site pocket is completely blocked by ImmD, which, by bringing a negatively charged cluster in opposition to a positively charged cluster on the surface of colicin D, appears to mimic the tRNA substrate backbone. Site-directed mutations affecting either the catalytic domain or the ImmD protein have led to the identification of the residues vital for catalytic activity and for the tight colicin D/ImmD interaction that inhibits colicin D toxicity and tRNase catalytic activity.
- LEBS, CNRS, UPR 9063, Gif sur Yvette, France.
Organizational Affiliation: