Distributed Computing and NMR Constraint-Based High-Resolution Structure Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal Folding
Takashima, H., Mimura, N., Ohkubo, T., Yoshida, T., Tamaoki, H., Kobayashi, Y.(2004) J Am Chem Soc 126: 4504-4505
- PubMed: 15070353
- DOI: https://doi.org/10.1021/ja031637w
- Primary Citation of Related Structures:
1V6R - PubMed Abstract:
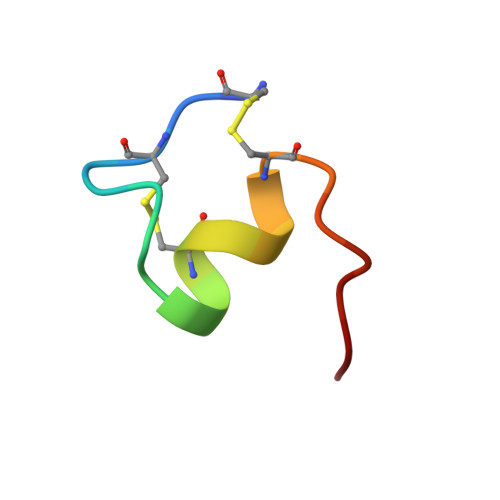
Distributed computing has been implemented to the solution structure determination of endothelin-1 to evaluate efficiency of the method for NMR constraint-based structure calculations. A key target of the investigation was determination of the C-terminal folding of the peptide, which had been dispersed in previous studies of NMR, despite its pharmacological significances. With use of tens of thousands of random initial structures to explore the conformational space comprehensively, we determined high-resolution structures with good convergences of C-terminal as well as previously defined N-terminal structures. The previous studies had missed the C-terminal convergence because of initial structure dependencies trapped in localized folding of the N-terminal region, which are strongly constricted by two disulfide bonds.
- Informatics and Knowledge Management at Novartis Institutes for BioMedical Research, Tsukuba Research Institute, Ohkubo 8, Tsukuba, Ibaraki, 300-2611 Japan. hiroyuki.takashima@pharma.novartis.com
Organizational Affiliation: