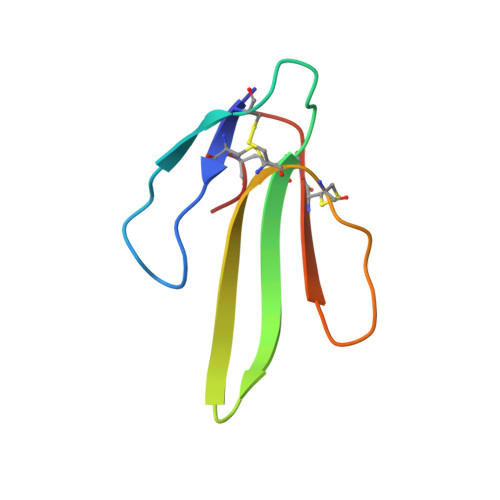
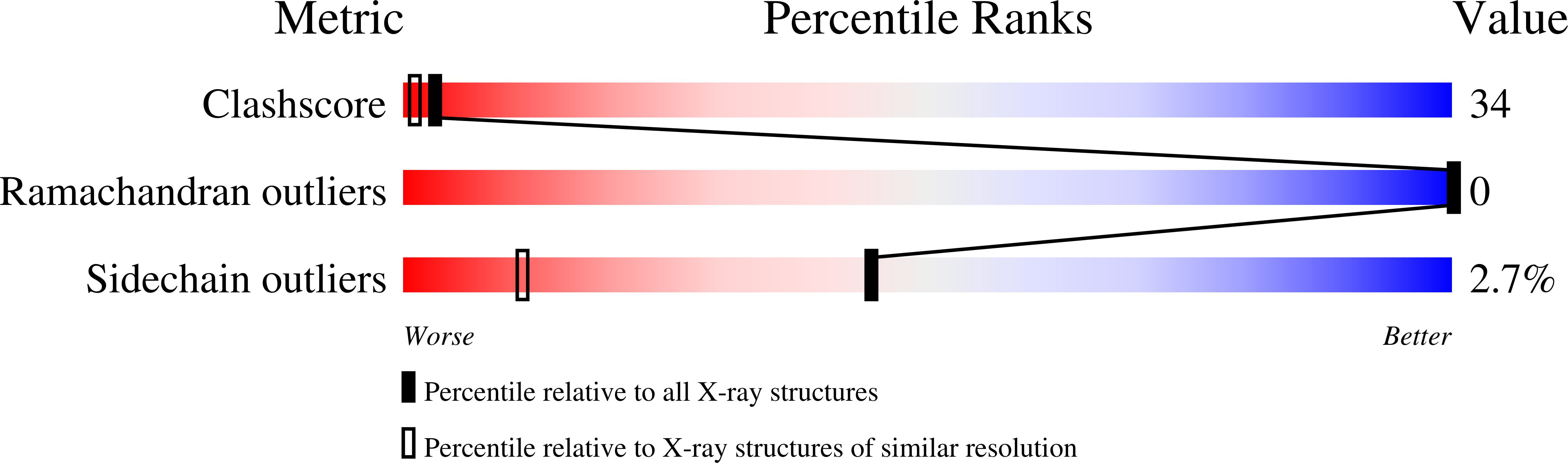The atomic resolution crystal structure of atratoxin determined by single wavelength anomalous diffraction phasing
Lou, X., Liu, Q., Tu, X., Wang, J., Teng, M., Niu, L., Schuller, D.J., Huang, Q., Hao, Q.(2004) J Biological Chem 279: 39094-39104
- PubMed: 15252034
- DOI: https://doi.org/10.1074/jbc.M403863200
- Primary Citation of Related Structures:
1V6P, 1VB0 - PubMed Abstract:
By using single wavelength anomalous diffraction phasing based on the anomalous signal from copper atoms, the crystal structure of atratoxin was determined at the resolution of 1.5 A and was refined to an ultrahigh resolution of 0.87 A. The ultrahigh resolution electron density maps allowed the modeling of 38 amino acid residues in alternate conformations and the location of 322 of 870 possible hydrogen atoms. To get accurate information at the atomic level, atratoxin-b (an analog of atratoxin with reduced toxicity) was also refined to an atomic resolution of 0.92 A. By the sequence and structural comparison of these two atratoxins, Arg(33) and Arg(36) were identified to be critical to their varied toxicity. The effect of copper ions on the distribution of hydrogen atoms in atratoxin was discussed, and the interactions between copper ions and protein residues were analyzed based on a statistical method, revealing a novel pentahedral copper-binding motif.
- Key Laboratory of Structural Biology, Chinese Academy of Sciences, University of Science and Technology of China, 96 Jinzhai Road, Hefei, Anhui 230026, China.
Organizational Affiliation: