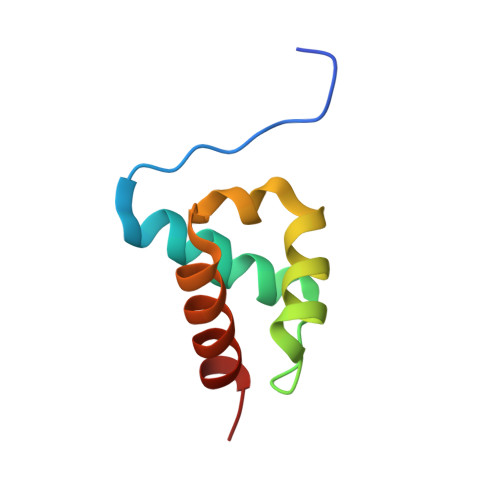
The Structure of an Ff Domain from Human Hypa/Fbp11.
Allen, M.D., Friedler, A., Schon, O., Bycroft, M.(2002) J Mol Biology 323: 411-416
- PubMed: 12381297
- DOI: https://doi.org/10.1016/s0022-2836(02)00968-3
- Primary Citation of Related Structures:
1UZC - PubMed Abstract:
The FF domain is a 60 amino acid residue phosphopeptide-binding module found in a variety of eukaryotic proteins including the transcription elongation factor CA150, the splicing factor Prp40 and p190RHOGAP. We have determined the structure of an FF domain from HYPA/FBP11. The domain is composed of three alpha helices arranged in an orthogonal bundle with a 3(10) helix in the loop between the second and third alpha helices. The structure differs from those of other phosphopeptide-binding domains and represents a novel phosphopeptide-binding fold.
- MRC Centre for Protein Engineering, Cambridge, UK.
Organizational Affiliation: